Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
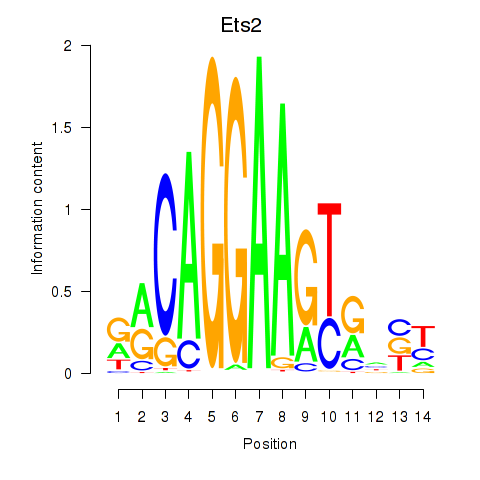
Results for Ets2
Z-value: 0.79
Transcription factors associated with Ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ets2
|
ENSMUSG00000022895.17 | E26 avian leukemia oncogene 2, 3' domain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ets2 | mm39_v1_chr16_+_95502911_95502960 | 0.73 | 1.6e-01 | Click! |
Activity profile of Ets2 motif
Sorted Z-values of Ets2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_70021155 | 0.59 |
ENSMUST00000041550.12
ENSMUST00000165951.8 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chr11_-_48707763 | 0.49 |
ENSMUST00000140800.2
|
Trim41
|
tripartite motif-containing 41 |
| chr9_+_110867807 | 0.48 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr3_-_90603013 | 0.45 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr11_-_55310724 | 0.37 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr11_-_48708159 | 0.35 |
ENSMUST00000047145.14
|
Trim41
|
tripartite motif-containing 41 |
| chr2_-_27974889 | 0.34 |
ENSMUST00000028179.15
ENSMUST00000117486.8 ENSMUST00000135472.2 |
Fcnb
|
ficolin B |
| chr1_+_40554513 | 0.32 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr8_+_73488496 | 0.30 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr3_+_89987749 | 0.29 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr11_-_6469494 | 0.26 |
ENSMUST00000134489.2
|
Myo1g
|
myosin IG |
| chr2_+_181023028 | 0.25 |
ENSMUST00000048077.12
|
Lime1
|
Lck interacting transmembrane adaptor 1 |
| chr1_+_40123858 | 0.25 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr1_+_85454323 | 0.24 |
ENSMUST00000239236.2
|
Gm7592
|
predicted gene 7592 |
| chr18_+_36916272 | 0.24 |
ENSMUST00000019287.9
|
Hars2
|
histidyl-tRNA synthetase 2 |
| chr6_+_52691204 | 0.24 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr9_-_119812042 | 0.24 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr10_-_40178182 | 0.23 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr17_+_26471889 | 0.22 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr8_+_39472981 | 0.21 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr11_+_74788904 | 0.21 |
ENSMUST00000045807.14
|
Tsr1
|
TSR1 20S rRNA accumulation |
| chr17_+_26471870 | 0.21 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chr11_-_118292678 | 0.20 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr12_-_55045887 | 0.20 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr17_+_34148868 | 0.20 |
ENSMUST00000173266.8
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_-_26063488 | 0.20 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr7_-_103928939 | 0.19 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr10_-_17898938 | 0.19 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr3_+_10077608 | 0.18 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chrX_+_55493325 | 0.18 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr10_+_80971054 | 0.18 |
ENSMUST00000125261.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chrX_-_149223883 | 0.18 |
ENSMUST00000163233.2
|
Tmem29
|
transmembrane protein 29 |
| chr9_+_107428713 | 0.17 |
ENSMUST00000093786.9
ENSMUST00000122225.8 |
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr19_-_4240984 | 0.17 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr2_+_32536594 | 0.17 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr17_-_12988492 | 0.17 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr8_+_80366247 | 0.17 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chr2_+_156681991 | 0.17 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr8_+_71261073 | 0.16 |
ENSMUST00000000808.8
ENSMUST00000212657.2 ENSMUST00000212146.2 |
Il12rb1
|
interleukin 12 receptor, beta 1 |
| chr8_+_86567600 | 0.16 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr4_+_149569717 | 0.16 |
ENSMUST00000030842.8
|
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr19_+_18690589 | 0.16 |
ENSMUST00000055792.8
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene |
| chr16_-_3690243 | 0.16 |
ENSMUST00000090522.5
|
Zfp597
|
zinc finger protein 597 |
| chr16_-_17906886 | 0.16 |
ENSMUST00000132241.2
ENSMUST00000139861.2 ENSMUST00000003620.13 |
Prodh
|
proline dehydrogenase |
| chrX_-_74460168 | 0.16 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr19_+_18690556 | 0.16 |
ENSMUST00000062753.3
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene |
| chr4_+_118266582 | 0.16 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr18_-_36916148 | 0.16 |
ENSMUST00000001416.8
|
Hars
|
histidyl-tRNA synthetase |
| chr11_+_120348919 | 0.16 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr8_-_122556219 | 0.15 |
ENSMUST00000174717.8
ENSMUST00000174192.2 |
Klhdc4
|
kelch domain containing 4 |
| chr5_-_121511476 | 0.15 |
ENSMUST00000202064.2
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr2_+_181023120 | 0.15 |
ENSMUST00000126611.8
|
Lime1
|
Lck interacting transmembrane adaptor 1 |
| chr9_-_44318597 | 0.15 |
ENSMUST00000217163.2
|
Trappc4
|
trafficking protein particle complex 4 |
| chr17_+_28491085 | 0.15 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr17_+_34128455 | 0.15 |
ENSMUST00000173626.8
ENSMUST00000170075.9 |
Daxx
|
Fas death domain-associated protein |
| chr3_-_130523954 | 0.15 |
ENSMUST00000196202.5
ENSMUST00000133802.6 ENSMUST00000062601.14 ENSMUST00000200517.2 |
Rpl34
|
ribosomal protein L34 |
| chr13_+_12410240 | 0.15 |
ENSMUST00000059270.10
|
Heatr1
|
HEAT repeat containing 1 |
| chr14_+_21881794 | 0.15 |
ENSMUST00000152562.8
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr11_-_20062876 | 0.15 |
ENSMUST00000000137.8
|
Actr2
|
ARP2 actin-related protein 2 |
| chr3_-_89325594 | 0.15 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr2_+_156681927 | 0.15 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr14_+_55120777 | 0.15 |
ENSMUST00000022806.10
|
Bcl2l2
|
BCL2-like 2 |
| chrX_-_106859842 | 0.15 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr19_-_40982576 | 0.14 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr1_-_16727133 | 0.14 |
ENSMUST00000185771.7
|
Eloc
|
elongin C |
| chr7_+_28533279 | 0.14 |
ENSMUST00000208971.2
ENSMUST00000066723.15 |
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr17_+_47908025 | 0.14 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr7_+_90091937 | 0.14 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chrX_-_133177638 | 0.14 |
ENSMUST00000113252.8
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr19_-_6002210 | 0.14 |
ENSMUST00000236013.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr5_+_149121458 | 0.14 |
ENSMUST00000122160.8
ENSMUST00000100410.10 ENSMUST00000119685.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr11_-_69214593 | 0.13 |
ENSMUST00000092973.6
|
Cntrob
|
centrobin, centrosomal BRCA2 interacting protein |
| chr14_+_55909816 | 0.13 |
ENSMUST00000227178.2
ENSMUST00000227914.2 |
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr11_+_70057449 | 0.13 |
ENSMUST00000102571.10
ENSMUST00000178945.8 ENSMUST00000000327.13 ENSMUST00000178567.3 |
Clec10a
|
C-type lectin domain family 10, member A |
| chrX_-_74460137 | 0.13 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr11_-_53321242 | 0.13 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr4_+_41760454 | 0.13 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr5_+_149121488 | 0.13 |
ENSMUST00000139474.8
ENSMUST00000117878.8 |
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr10_-_83484576 | 0.13 |
ENSMUST00000020500.14
|
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr14_+_55120875 | 0.13 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr7_+_141040988 | 0.13 |
ENSMUST00000053670.12
|
Cracr2b
|
calcium release activated channel regulator 2B |
| chr17_+_29579882 | 0.13 |
ENSMUST00000024810.8
|
Fgd2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr10_-_115220293 | 0.13 |
ENSMUST00000020346.6
ENSMUST00000218842.2 ENSMUST00000218989.2 |
Thap2
|
THAP domain containing, apoptosis associated protein 2 |
| chr3_-_33898405 | 0.13 |
ENSMUST00000029222.8
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr3_+_87754310 | 0.13 |
ENSMUST00000029709.7
|
Sh2d2a
|
SH2 domain containing 2A |
| chr12_+_80509869 | 0.13 |
ENSMUST00000038185.10
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr7_-_45116197 | 0.13 |
ENSMUST00000211195.2
ENSMUST00000210019.2 |
Bax
|
BCL2-associated X protein |
| chr5_+_34700709 | 0.13 |
ENSMUST00000101316.10
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr17_-_26063391 | 0.13 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr9_+_21504018 | 0.12 |
ENSMUST00000062125.11
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr8_+_123980934 | 0.12 |
ENSMUST00000001092.15
|
Zfp276
|
zinc finger protein (C2H2 type) 276 |
| chr19_-_4241034 | 0.12 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr3_+_157239988 | 0.12 |
ENSMUST00000029831.16
ENSMUST00000106057.8 |
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr14_-_32907023 | 0.12 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr18_-_84700170 | 0.12 |
ENSMUST00000168419.3
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr7_+_79919380 | 0.12 |
ENSMUST00000032749.12
|
Vps33b
|
vacuolar protein sorting 33B |
| chr11_+_58868919 | 0.12 |
ENSMUST00000108809.8
ENSMUST00000108810.10 ENSMUST00000093061.7 |
Trim11
|
tripartite motif-containing 11 |
| chr7_-_104114384 | 0.12 |
ENSMUST00000076922.6
|
Trim30a
|
tripartite motif-containing 30A |
| chr1_-_121255448 | 0.12 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr8_+_111573646 | 0.12 |
ENSMUST00000172668.8
ENSMUST00000034203.17 ENSMUST00000174398.8 |
Cog4
|
component of oligomeric golgi complex 4 |
| chr4_-_118266416 | 0.12 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr11_+_86574811 | 0.12 |
ENSMUST00000108022.8
ENSMUST00000108021.2 |
Ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr2_+_24276616 | 0.12 |
ENSMUST00000166388.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr9_+_63509925 | 0.12 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr15_+_8138805 | 0.12 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr3_+_129326004 | 0.12 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr3_+_94600863 | 0.12 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr11_-_97040858 | 0.12 |
ENSMUST00000118375.8
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr9_-_60595401 | 0.12 |
ENSMUST00000114034.9
ENSMUST00000065603.12 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr7_-_79882228 | 0.11 |
ENSMUST00000123279.8
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr5_+_149121355 | 0.11 |
ENSMUST00000050472.16
|
Uspl1
|
ubiquitin specific peptidase like 1 |
| chr8_-_85807281 | 0.11 |
ENSMUST00000152785.8
|
Wdr83
|
WD repeat domain containing 83 |
| chr2_+_3425159 | 0.11 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr2_+_38898065 | 0.11 |
ENSMUST00000112862.7
|
Arpc5l
|
actin related protein 2/3 complex, subunit 5-like |
| chr1_-_132953068 | 0.11 |
ENSMUST00000186617.7
ENSMUST00000067429.10 ENSMUST00000067398.13 ENSMUST00000188090.7 |
Mdm4
|
transformed mouse 3T3 cell double minute 4 |
| chr19_+_8748347 | 0.11 |
ENSMUST00000010248.4
|
Tmem223
|
transmembrane protein 223 |
| chr12_+_17398421 | 0.11 |
ENSMUST00000046011.12
|
Nol10
|
nucleolar protein 10 |
| chr16_-_10994135 | 0.11 |
ENSMUST00000037633.16
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr6_+_88442391 | 0.11 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr10_-_79369584 | 0.11 |
ENSMUST00000218241.2
ENSMUST00000166804.2 ENSMUST00000063879.13 |
Plpp2
|
phospholipid phosphatase 2 |
| chr7_-_45116216 | 0.11 |
ENSMUST00000210392.2
ENSMUST00000211365.2 |
Bax
|
BCL2-associated X protein |
| chr8_+_85786684 | 0.10 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr17_-_25155868 | 0.10 |
ENSMUST00000115228.9
ENSMUST00000117509.8 ENSMUST00000121723.8 ENSMUST00000119115.8 ENSMUST00000121787.8 ENSMUST00000088345.12 ENSMUST00000120035.8 ENSMUST00000115229.10 ENSMUST00000178969.8 |
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr15_-_75801630 | 0.10 |
ENSMUST00000229289.2
ENSMUST00000229641.2 |
Gfus
|
GDP-L-fucose synthase |
| chr8_+_22966736 | 0.10 |
ENSMUST00000067786.9
|
Slc20a2
|
solute carrier family 20, member 2 |
| chr2_-_91480096 | 0.10 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr16_-_38533597 | 0.10 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr14_-_75991903 | 0.10 |
ENSMUST00000049168.9
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr4_+_3574855 | 0.10 |
ENSMUST00000052712.6
|
Tgs1
|
trimethylguanosine synthase 1 |
| chr4_+_43562706 | 0.10 |
ENSMUST00000167751.2
ENSMUST00000132631.2 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr4_+_149569672 | 0.10 |
ENSMUST00000124413.8
ENSMUST00000141293.8 |
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr6_+_125048230 | 0.10 |
ENSMUST00000140346.9
ENSMUST00000171989.3 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr5_-_35886605 | 0.10 |
ENSMUST00000070203.14
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr1_+_180678677 | 0.10 |
ENSMUST00000038091.8
|
Sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr9_-_66421868 | 0.10 |
ENSMUST00000056890.10
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr14_+_8348779 | 0.10 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr1_-_170133901 | 0.10 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr17_+_34148485 | 0.10 |
ENSMUST00000047503.16
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr6_+_54249817 | 0.10 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr12_-_13299136 | 0.10 |
ENSMUST00000221623.2
|
Ddx1
|
DEAD box helicase 1 |
| chr11_-_102208615 | 0.10 |
ENSMUST00000107117.9
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr7_-_127805518 | 0.10 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr7_-_121732067 | 0.10 |
ENSMUST00000106469.8
ENSMUST00000063587.13 ENSMUST00000106468.7 ENSMUST00000098068.10 |
Palb2
|
partner and localizer of BRCA2 |
| chr9_+_45818250 | 0.09 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr7_+_101043568 | 0.09 |
ENSMUST00000098243.4
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_+_87683592 | 0.09 |
ENSMUST00000144047.8
ENSMUST00000027512.13 ENSMUST00000113186.8 ENSMUST00000113190.3 |
Atg16l1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr3_+_157239740 | 0.09 |
ENSMUST00000106058.8
|
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr9_-_65815958 | 0.09 |
ENSMUST00000119245.8
ENSMUST00000134338.8 ENSMUST00000179395.8 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr17_+_46961250 | 0.09 |
ENSMUST00000043464.14
|
Cul7
|
cullin 7 |
| chr2_+_43638814 | 0.09 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr19_+_32463151 | 0.09 |
ENSMUST00000025827.10
|
Minpp1
|
multiple inositol polyphosphate histidine phosphatase 1 |
| chr11_-_102208449 | 0.09 |
ENSMUST00000107123.10
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr9_+_65253374 | 0.09 |
ENSMUST00000048184.4
ENSMUST00000214433.2 |
Pdcd7
|
programmed cell death 7 |
| chr15_-_100449668 | 0.09 |
ENSMUST00000229581.2
ENSMUST00000231138.2 |
Tfcp2
|
transcription factor CP2 |
| chr7_-_79882313 | 0.09 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr7_-_140680437 | 0.09 |
ENSMUST00000210167.2
ENSMUST00000209294.2 ENSMUST00000097958.3 |
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr19_-_38212544 | 0.09 |
ENSMUST00000067167.6
|
Fra10ac1
|
FRA10AC1 homolog (human) |
| chr2_+_31462659 | 0.09 |
ENSMUST00000113482.8
|
Fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr1_-_16727242 | 0.09 |
ENSMUST00000186948.7
ENSMUST00000187910.7 ENSMUST00000115352.10 |
Eloc
|
elongin C |
| chr11_-_23448030 | 0.09 |
ENSMUST00000020529.13
|
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr12_+_76353835 | 0.09 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr1_-_93659622 | 0.09 |
ENSMUST00000189728.7
|
Thap4
|
THAP domain containing 4 |
| chr8_-_85807308 | 0.09 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr12_-_4283926 | 0.09 |
ENSMUST00000111169.10
ENSMUST00000020981.12 |
Cenpo
|
centromere protein O |
| chr10_+_4561974 | 0.09 |
ENSMUST00000105590.8
ENSMUST00000067086.14 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr19_-_46950355 | 0.09 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr9_-_35122261 | 0.09 |
ENSMUST00000043805.15
ENSMUST00000142595.8 ENSMUST00000127996.8 |
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr11_-_5492175 | 0.09 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr11_+_97253221 | 0.09 |
ENSMUST00000238729.2
ENSMUST00000045540.4 |
Socs7
|
suppressor of cytokine signaling 7 |
| chr9_+_7184514 | 0.09 |
ENSMUST00000215683.2
ENSMUST00000034499.10 |
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr19_+_8931187 | 0.09 |
ENSMUST00000096239.7
|
Tut1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr17_-_3608056 | 0.09 |
ENSMUST00000041003.8
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr16_-_18630722 | 0.09 |
ENSMUST00000000028.14
ENSMUST00000115585.2 |
Cdc45
|
cell division cycle 45 |
| chr2_+_120294046 | 0.09 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr10_-_17898838 | 0.09 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr14_+_79086665 | 0.09 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr9_-_44318823 | 0.09 |
ENSMUST00000034623.8
|
Trappc4
|
trafficking protein particle complex 4 |
| chr17_-_29483075 | 0.08 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr4_+_59805829 | 0.08 |
ENSMUST00000030080.7
|
Snx30
|
sorting nexin family member 30 |
| chr15_-_81244940 | 0.08 |
ENSMUST00000023040.9
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17 |
| chr15_-_75801575 | 0.08 |
ENSMUST00000229951.2
ENSMUST00000023231.7 ENSMUST00000230736.2 |
Gfus
|
GDP-L-fucose synthase |
| chr4_-_116485118 | 0.08 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr13_-_58276353 | 0.08 |
ENSMUST00000007980.7
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr8_-_65302573 | 0.08 |
ENSMUST00000210166.2
|
Klhl2
|
kelch-like 2, Mayven |
| chr19_+_18609291 | 0.08 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr8_-_111573401 | 0.08 |
ENSMUST00000042012.7
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr15_+_84926909 | 0.08 |
ENSMUST00000229203.2
|
Fam118a
|
family with sequence similarity 118, member A |
| chr6_+_137712076 | 0.08 |
ENSMUST00000064910.7
|
Strap
|
serine/threonine kinase receptor associated protein |
| chr9_+_107445101 | 0.08 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr12_+_117621644 | 0.08 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chrX_-_133177717 | 0.08 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr1_-_121255400 | 0.08 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr6_+_108190163 | 0.08 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr11_+_5905693 | 0.08 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr16_-_10993919 | 0.08 |
ENSMUST00000142389.3
ENSMUST00000138185.9 |
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr2_+_72128239 | 0.08 |
ENSMUST00000144111.2
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr13_-_23553327 | 0.08 |
ENSMUST00000125328.2
ENSMUST00000145451.8 ENSMUST00000050101.9 |
Zfp322a
|
zinc finger protein 322A |
| chr2_+_113271409 | 0.08 |
ENSMUST00000081349.9
|
Fmn1
|
formin 1 |
| chr10_+_61133549 | 0.08 |
ENSMUST00000219375.2
|
Prf1
|
perforin 1 (pore forming protein) |
| chr7_-_125968653 | 0.08 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ets2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.4 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.5 | GO:0051673 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.4 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.3 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.1 | 0.2 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.2 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:1905072 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.4 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.2 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0032262 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.0 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.0 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.3 | GO:0097144 | BAX complex(GO:0097144) |
| 0.1 | 0.5 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.4 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.1 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.1 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |