Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
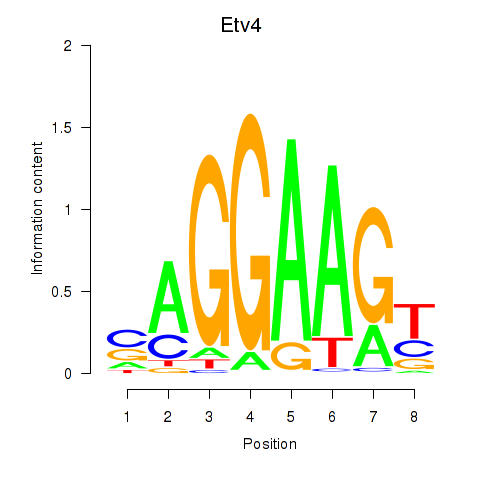
Results for Etv4
Z-value: 1.32
Transcription factors associated with Etv4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv4
|
ENSMUSG00000017724.15 | ets variant 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv4 | mm39_v1_chr11_-_101676076_101676197 | -0.12 | 8.5e-01 | Click! |
Activity profile of Etv4 motif
Sorted Z-values of Etv4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_110305705 | 0.84 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chr8_-_106052884 | 0.81 |
ENSMUST00000210412.2
ENSMUST00000210801.2 ENSMUST00000070508.8 |
Lrrc29
|
leucine rich repeat containing 29 |
| chr6_-_100648086 | 0.71 |
ENSMUST00000089245.7
ENSMUST00000113312.9 ENSMUST00000170667.8 |
Shq1
|
SHQ1 homolog (S. cerevisiae) |
| chr11_-_48708159 | 0.68 |
ENSMUST00000047145.14
|
Trim41
|
tripartite motif-containing 41 |
| chr17_-_51486196 | 0.66 |
ENSMUST00000024717.10
ENSMUST00000224528.2 |
Tbc1d5
|
TBC1 domain family, member 5 |
| chr9_+_110306052 | 0.62 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chr10_-_12689345 | 0.61 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr13_-_32967937 | 0.57 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr15_+_6609322 | 0.54 |
ENSMUST00000090461.12
|
Fyb
|
FYN binding protein |
| chr18_-_84607615 | 0.52 |
ENSMUST00000125763.3
|
Zfp407
|
zinc finger protein 407 |
| chr19_-_53933052 | 0.51 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr4_-_156312961 | 0.50 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr10_-_85847697 | 0.49 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr2_-_101627999 | 0.47 |
ENSMUST00000171088.8
ENSMUST00000043845.14 |
Prr5l
|
proline rich 5 like |
| chr1_-_173569301 | 0.46 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr16_-_38533597 | 0.45 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr9_-_110237276 | 0.45 |
ENSMUST00000040021.12
|
Ptpn23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr11_+_70538083 | 0.44 |
ENSMUST00000037534.8
|
Rnf167
|
ring finger protein 167 |
| chr11_-_4110286 | 0.44 |
ENSMUST00000093381.11
ENSMUST00000101626.9 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr10_-_126877382 | 0.44 |
ENSMUST00000116231.4
|
Eef1akmt3
|
EEF1A lysine methyltransferase 3 |
| chr15_-_98465516 | 0.43 |
ENSMUST00000012104.7
|
Ccnt1
|
cyclin T1 |
| chr2_+_11647610 | 0.43 |
ENSMUST00000028111.6
|
Il2ra
|
interleukin 2 receptor, alpha chain |
| chr2_+_113271409 | 0.42 |
ENSMUST00000081349.9
|
Fmn1
|
formin 1 |
| chr3_+_137573436 | 0.42 |
ENSMUST00000090178.10
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr11_-_51647204 | 0.42 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr1_+_16758629 | 0.42 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr11_-_95733235 | 0.41 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr10_+_80633865 | 0.40 |
ENSMUST00000148665.8
|
Sf3a2
|
splicing factor 3a, subunit 2 |
| chr19_+_10502679 | 0.40 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr5_-_110596387 | 0.40 |
ENSMUST00000198768.3
|
Fbrsl1
|
fibrosin-like 1 |
| chr8_-_123768759 | 0.40 |
ENSMUST00000098334.13
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr19_+_10502612 | 0.39 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr5_-_25305621 | 0.39 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr5_-_135494775 | 0.39 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr17_-_16046780 | 0.37 |
ENSMUST00000232638.2
ENSMUST00000170578.3 |
Rgmb
|
repulsive guidance molecule family member B |
| chr5_+_29639662 | 0.37 |
ENSMUST00000001611.11
|
Nom1
|
nucleolar protein with MIF4G domain 1 |
| chr6_-_97408367 | 0.37 |
ENSMUST00000124050.3
|
Frmd4b
|
FERM domain containing 4B |
| chr2_-_91480096 | 0.37 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr1_-_160134873 | 0.37 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr2_-_167334746 | 0.37 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chr7_+_126365506 | 0.36 |
ENSMUST00000032944.9
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr5_-_110596352 | 0.36 |
ENSMUST00000069483.12
ENSMUST00000200293.2 |
Fbrsl1
|
fibrosin-like 1 |
| chr12_+_76580386 | 0.35 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr12_-_31549538 | 0.35 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr2_+_164897547 | 0.35 |
ENSMUST00000017799.12
ENSMUST00000073707.9 |
Cd40
|
CD40 antigen |
| chr3_-_122778052 | 0.35 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr15_+_30173197 | 0.35 |
ENSMUST00000226119.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr12_+_76593799 | 0.34 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr13_-_17979675 | 0.34 |
ENSMUST00000223490.2
|
Cdk13
|
cyclin-dependent kinase 13 |
| chr15_-_76079891 | 0.34 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr19_+_18609291 | 0.34 |
ENSMUST00000042392.14
ENSMUST00000237347.2 |
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr4_-_123458465 | 0.33 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_-_89951611 | 0.33 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr6_-_83433357 | 0.33 |
ENSMUST00000186548.7
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr12_-_84265609 | 0.33 |
ENSMUST00000046266.13
ENSMUST00000220974.2 |
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr2_+_3425159 | 0.33 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr4_-_63321591 | 0.33 |
ENSMUST00000035724.5
|
Akna
|
AT-hook transcription factor |
| chr8_-_123768984 | 0.33 |
ENSMUST00000212937.2
|
Ankrd11
|
ankyrin repeat domain 11 |
| chr13_-_98951627 | 0.33 |
ENSMUST00000224992.2
ENSMUST00000225840.2 |
Fcho2
|
FCH domain only 2 |
| chr9_-_43027809 | 0.33 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr16_+_14523696 | 0.32 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr17_-_80597495 | 0.32 |
ENSMUST00000086555.11
|
Dhx57
|
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 |
| chr11_+_54413772 | 0.32 |
ENSMUST00000207429.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr19_-_10502468 | 0.31 |
ENSMUST00000025570.8
ENSMUST00000236455.2 |
Sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr10_+_7708178 | 0.31 |
ENSMUST00000039484.6
|
Zc3h12d
|
zinc finger CCCH type containing 12D |
| chr17_-_59320257 | 0.31 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr12_-_65012270 | 0.31 |
ENSMUST00000222508.2
|
Klhl28
|
kelch-like 28 |
| chr14_+_51366512 | 0.30 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr11_+_46345747 | 0.30 |
ENSMUST00000020668.15
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr15_+_76361604 | 0.30 |
ENSMUST00000226872.2
ENSMUST00000072838.6 ENSMUST00000227478.2 ENSMUST00000228371.2 ENSMUST00000229363.2 |
Hsf1
|
heat shock factor 1 |
| chr7_+_130375799 | 0.30 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr9_-_57743989 | 0.30 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr1_+_131936022 | 0.29 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr4_+_129229373 | 0.29 |
ENSMUST00000141235.8
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr15_-_101268036 | 0.29 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr4_-_156312996 | 0.29 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr13_+_20369377 | 0.29 |
ENSMUST00000221067.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr14_-_32907446 | 0.29 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr14_-_19261196 | 0.29 |
ENSMUST00000112797.12
ENSMUST00000225885.2 |
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr13_+_41040657 | 0.29 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr2_+_145627900 | 0.29 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr1_-_121255448 | 0.28 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr1_+_171668122 | 0.28 |
ENSMUST00000135386.2
|
Cd84
|
CD84 antigen |
| chr17_+_35354148 | 0.28 |
ENSMUST00000166426.9
ENSMUST00000025250.14 |
Bag6
|
BCL2-associated athanogene 6 |
| chr11_+_117545037 | 0.28 |
ENSMUST00000026658.13
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr10_-_95159933 | 0.28 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr11_+_54413698 | 0.28 |
ENSMUST00000108895.8
ENSMUST00000101206.10 |
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr11_+_98851238 | 0.28 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr5_-_90514360 | 0.28 |
ENSMUST00000168058.7
ENSMUST00000014421.15 |
Ankrd17
|
ankyrin repeat domain 17 |
| chr10_-_83484576 | 0.28 |
ENSMUST00000020500.14
|
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr10_-_5773881 | 0.27 |
ENSMUST00000019908.9
|
Mtrf1l
|
mitochondrial translational release factor 1-like |
| chr1_-_123973223 | 0.27 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr3_+_96484294 | 0.27 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr15_+_30172716 | 0.27 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr2_+_73142945 | 0.27 |
ENSMUST00000090811.11
ENSMUST00000112050.2 |
Scrn3
|
secernin 3 |
| chr11_-_103254257 | 0.27 |
ENSMUST00000092557.6
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr15_-_9529898 | 0.27 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr7_+_127475968 | 0.26 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr11_+_70431063 | 0.26 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr2_-_180351778 | 0.26 |
ENSMUST00000103057.8
ENSMUST00000103055.8 |
Dido1
|
death inducer-obliterator 1 |
| chr9_+_57496725 | 0.26 |
ENSMUST00000053230.7
|
Ulk3
|
unc-51-like kinase 3 |
| chr10_-_83484467 | 0.26 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr5_-_90514061 | 0.25 |
ENSMUST00000081914.13
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr19_-_40982576 | 0.25 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr12_+_102249375 | 0.25 |
ENSMUST00000101114.11
ENSMUST00000150795.8 |
Rin3
|
Ras and Rab interactor 3 |
| chr4_-_135221810 | 0.25 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr16_+_24212284 | 0.25 |
ENSMUST00000038053.14
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_113548201 | 0.25 |
ENSMUST00000148736.8
ENSMUST00000142069.8 ENSMUST00000134418.8 |
Cog1
|
component of oligomeric golgi complex 1 |
| chr5_+_143803540 | 0.25 |
ENSMUST00000100487.6
|
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr5_+_129578285 | 0.25 |
ENSMUST00000053737.9
|
Sfswap
|
splicing factor SWAP |
| chr1_-_39517761 | 0.25 |
ENSMUST00000193823.2
ENSMUST00000054462.11 |
Tbc1d8
|
TBC1 domain family, member 8 |
| chr4_-_62278761 | 0.25 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr2_+_48839505 | 0.24 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr7_+_79919380 | 0.24 |
ENSMUST00000032749.12
|
Vps33b
|
vacuolar protein sorting 33B |
| chr3_-_90603013 | 0.24 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr7_+_126184108 | 0.24 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr5_+_23992689 | 0.24 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr7_+_139616298 | 0.24 |
ENSMUST00000168194.3
ENSMUST00000210882.2 |
Zfp511
|
zinc finger protein 511 |
| chr11_+_54413673 | 0.24 |
ENSMUST00000102743.10
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr8_-_46452896 | 0.23 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr17_+_34251041 | 0.23 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta |
| chr3_+_105811712 | 0.23 |
ENSMUST00000000574.3
|
Adora3
|
adenosine A3 receptor |
| chr1_+_138891155 | 0.23 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr9_+_106099797 | 0.23 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr11_-_48707763 | 0.23 |
ENSMUST00000140800.2
|
Trim41
|
tripartite motif-containing 41 |
| chr17_-_32503107 | 0.23 |
ENSMUST00000237692.2
|
Brd4
|
bromodomain containing 4 |
| chr2_-_73284262 | 0.23 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr8_+_3403415 | 0.23 |
ENSMUST00000098966.5
ENSMUST00000239102.2 |
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr13_+_109397184 | 0.23 |
ENSMUST00000153234.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_+_125529911 | 0.23 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr16_-_16417842 | 0.23 |
ENSMUST00000162671.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr15_-_88703482 | 0.23 |
ENSMUST00000043087.15
ENSMUST00000162183.8 |
Alg12
|
asparagine-linked glycosylation 12 (alpha-1,6-mannosyltransferase) |
| chrX_+_109857866 | 0.23 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr17_-_32503060 | 0.22 |
ENSMUST00000003726.16
ENSMUST00000121285.8 ENSMUST00000120276.9 |
Brd4
|
bromodomain containing 4 |
| chr5_+_34700709 | 0.22 |
ENSMUST00000101316.10
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chrX_-_74128363 | 0.22 |
ENSMUST00000114104.2
ENSMUST00000114109.8 ENSMUST00000037374.11 |
Gab3
|
growth factor receptor bound protein 2-associated protein 3 |
| chr3_+_137849239 | 0.22 |
ENSMUST00000162864.8
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr5_-_90514034 | 0.22 |
ENSMUST00000218526.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr15_-_101262452 | 0.22 |
ENSMUST00000230909.2
|
Krt80
|
keratin 80 |
| chr16_-_4698148 | 0.22 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr16_-_3794404 | 0.22 |
ENSMUST00000180200.8
|
Nlrc3
|
NLR family, CARD domain containing 3 |
| chr14_-_55909314 | 0.22 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr1_+_131935342 | 0.22 |
ENSMUST00000086556.12
|
Elk4
|
ELK4, member of ETS oncogene family |
| chrX_-_12628309 | 0.22 |
ENSMUST00000096495.11
ENSMUST00000076016.6 |
Med14
|
mediator complex subunit 14 |
| chr8_-_73228953 | 0.22 |
ENSMUST00000079510.6
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr19_+_18609343 | 0.22 |
ENSMUST00000159572.9
|
Nmrk1
|
nicotinamide riboside kinase 1 |
| chr2_-_27317004 | 0.22 |
ENSMUST00000056176.8
|
Vav2
|
vav 2 oncogene |
| chr6_-_99005212 | 0.22 |
ENSMUST00000177437.8
ENSMUST00000177229.8 ENSMUST00000124058.8 |
Foxp1
|
forkhead box P1 |
| chr5_+_33021042 | 0.22 |
ENSMUST00000149350.8
ENSMUST00000118698.8 ENSMUST00000150130.8 ENSMUST00000049780.13 ENSMUST00000087897.11 ENSMUST00000119705.8 ENSMUST00000125574.8 |
Depdc5
|
DEP domain containing 5 |
| chr11_-_51647290 | 0.22 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr5_-_31684036 | 0.22 |
ENSMUST00000202421.2
ENSMUST00000201769.4 ENSMUST00000065388.11 |
Supt7l
|
SPT7-like, STAGA complex gamma subunit |
| chr17_+_35454833 | 0.21 |
ENSMUST00000118384.8
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr4_-_62278673 | 0.21 |
ENSMUST00000084527.10
ENSMUST00000098033.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr3_-_89820451 | 0.21 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr9_+_108708939 | 0.21 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr14_-_75185281 | 0.21 |
ENSMUST00000088970.7
ENSMUST00000228252.2 |
Lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr8_-_73229056 | 0.21 |
ENSMUST00000212991.2
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr7_-_132415257 | 0.21 |
ENSMUST00000097999.9
|
Fam53b
|
family with sequence similarity 53, member B |
| chr11_+_103061905 | 0.21 |
ENSMUST00000042286.12
ENSMUST00000218163.2 |
Fmnl1
|
formin-like 1 |
| chr3_+_60380243 | 0.21 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr4_+_62581857 | 0.21 |
ENSMUST00000126338.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr3_-_88277037 | 0.21 |
ENSMUST00000075523.11
|
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr11_-_62348115 | 0.21 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr6_+_134958681 | 0.20 |
ENSMUST00000167323.3
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr3_+_137849189 | 0.20 |
ENSMUST00000040321.13
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr9_+_57818243 | 0.20 |
ENSMUST00000216925.2
ENSMUST00000163329.2 ENSMUST00000213654.2 ENSMUST00000217132.2 ENSMUST00000216841.2 ENSMUST00000214086.2 |
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chrX_+_162691978 | 0.20 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr11_+_75404356 | 0.20 |
ENSMUST00000042808.13
ENSMUST00000118243.2 |
Scarf1
|
scavenger receptor class F, member 1 |
| chr1_+_172204109 | 0.20 |
ENSMUST00000052455.4
|
Pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr11_+_46345784 | 0.20 |
ENSMUST00000109229.2
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr11_-_79414542 | 0.20 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chr2_+_61423469 | 0.20 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr4_-_116413092 | 0.20 |
ENSMUST00000069674.6
ENSMUST00000106478.9 |
Tmem69
|
transmembrane protein 69 |
| chr11_-_44361289 | 0.20 |
ENSMUST00000102795.4
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr4_-_118266416 | 0.20 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr11_+_77873535 | 0.20 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr17_+_35354172 | 0.20 |
ENSMUST00000172571.8
ENSMUST00000173491.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr15_+_81511486 | 0.20 |
ENSMUST00000206833.2
|
Ep300
|
E1A binding protein p300 |
| chr1_-_133728779 | 0.20 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_+_117545618 | 0.20 |
ENSMUST00000106344.8
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr2_-_58990967 | 0.19 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr14_+_20732804 | 0.19 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr11_+_60668350 | 0.19 |
ENSMUST00000102667.5
ENSMUST00000056907.7 |
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 homolog (human) |
| chr8_+_123980934 | 0.19 |
ENSMUST00000001092.15
|
Zfp276
|
zinc finger protein (C2H2 type) 276 |
| chr13_+_41013230 | 0.19 |
ENSMUST00000110191.10
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_-_75345482 | 0.19 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chr17_+_35354655 | 0.19 |
ENSMUST00000174478.8
ENSMUST00000174281.9 ENSMUST00000173550.8 |
Bag6
|
BCL2-associated athanogene 6 |
| chr5_-_142891686 | 0.19 |
ENSMUST00000106216.3
|
Actb
|
actin, beta |
| chr9_+_86349226 | 0.19 |
ENSMUST00000188675.7
|
Dop1a
|
DOP1 leucine zipper like protein A |
| chr12_-_84240781 | 0.19 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr2_-_102231208 | 0.19 |
ENSMUST00000102573.8
|
Trim44
|
tripartite motif-containing 44 |
| chr14_-_122153185 | 0.19 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18 |
| chr2_+_91480513 | 0.19 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr9_-_66033841 | 0.19 |
ENSMUST00000137542.2
|
Snx1
|
sorting nexin 1 |
| chr10_-_128245501 | 0.19 |
ENSMUST00000172348.8
ENSMUST00000166608.8 ENSMUST00000164199.8 ENSMUST00000171370.2 ENSMUST00000026439.14 |
Nabp2
|
nucleic acid binding protein 2 |
| chr12_+_102249294 | 0.19 |
ENSMUST00000056950.14
|
Rin3
|
Ras and Rab interactor 3 |
| chr13_+_84370405 | 0.18 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr13_+_3684032 | 0.18 |
ENSMUST00000042288.8
|
Asb13
|
ankyrin repeat and SOCS box-containing 13 |
| chr2_-_168072295 | 0.18 |
ENSMUST00000154111.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr7_-_126294902 | 0.18 |
ENSMUST00000144897.2
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.5 | GO:0002859 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.5 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.8 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.6 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.6 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.1 | 0.2 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.3 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.2 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.1 | 0.2 | GO:0045763 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.2 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.1 | 0.2 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.2 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.2 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.4 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.4 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.5 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.2 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.5 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1905076 | regulation of interleukin-17 secretion(GO:1905076) |
| 0.0 | 0.1 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.8 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.3 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.3 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.2 | GO:0051532 | regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.1 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 0.6 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.6 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:1904306 | positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 0.2 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.0 | GO:0090076 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.1 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.0 | 0.5 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.2 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:1904093 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.0 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 0.3 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.3 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.7 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.4 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.3 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.4 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 0.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.2 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.3 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.2 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.1 | 1.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0004615 | phosphomannomutase activity(GO:0004615) |
| 0.0 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |