Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
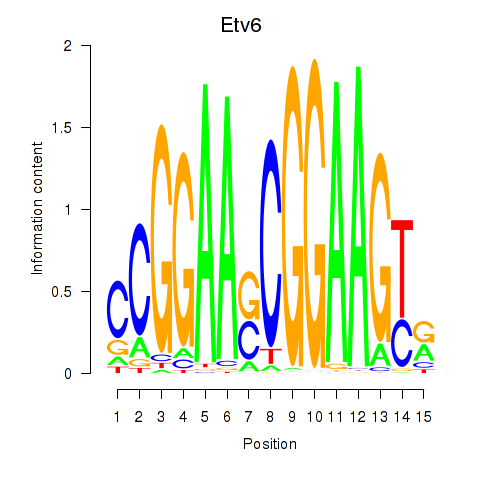
Results for Etv6
Z-value: 1.62
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSMUSG00000030199.17 | ets variant 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv6 | mm39_v1_chr6_+_134012602_134012683 | -0.70 | 1.9e-01 | Click! |
Activity profile of Etv6 motif
Sorted Z-values of Etv6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_94794256 | 0.83 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr2_+_130266253 | 0.72 |
ENSMUST00000128994.8
ENSMUST00000028900.11 |
Vps16
|
VSP16 CORVET/HOPS core subunit |
| chr1_-_133537953 | 0.65 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr8_-_73324877 | 0.58 |
ENSMUST00000058733.9
ENSMUST00000167290.8 |
Smim7
|
small integral membrane protein 7 |
| chr13_+_4241149 | 0.57 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr11_-_35725317 | 0.52 |
ENSMUST00000018992.4
|
Rars
|
arginyl-tRNA synthetase |
| chr7_+_18883647 | 0.50 |
ENSMUST00000049294.4
|
Snrpd2
|
small nuclear ribonucleoprotein D2 |
| chr4_+_118478357 | 0.48 |
ENSMUST00000147373.2
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr11_+_58221538 | 0.47 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr13_-_58276353 | 0.47 |
ENSMUST00000007980.7
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr2_+_179899166 | 0.47 |
ENSMUST00000059080.7
|
Rps21
|
ribosomal protein S21 |
| chr9_+_46184362 | 0.46 |
ENSMUST00000156440.8
ENSMUST00000114552.4 |
Zpr1
|
ZPR1 zinc finger |
| chr2_-_90735171 | 0.45 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr15_-_85695855 | 0.45 |
ENSMUST00000134631.8
ENSMUST00000154814.2 ENSMUST00000071876.13 ENSMUST00000150995.8 |
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chrX_-_168103266 | 0.45 |
ENSMUST00000033717.9
ENSMUST00000112115.2 |
Hccs
|
holocytochrome c synthetase |
| chr6_-_120799641 | 0.45 |
ENSMUST00000205049.3
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr7_-_45083688 | 0.45 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr19_+_8919228 | 0.43 |
ENSMUST00000096240.3
|
Mta2
|
metastasis-associated gene family, member 2 |
| chr14_-_30723549 | 0.43 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr6_-_113717689 | 0.42 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr15_-_57755753 | 0.41 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr8_+_85583611 | 0.41 |
ENSMUST00000003906.13
ENSMUST00000109754.2 |
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chrX_-_133442596 | 0.40 |
ENSMUST00000054213.5
|
Timm8a1
|
translocase of inner mitochondrial membrane 8A1 |
| chr9_-_78396407 | 0.40 |
ENSMUST00000154207.8
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_+_58221569 | 0.40 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr10_+_130158737 | 0.39 |
ENSMUST00000217702.2
ENSMUST00000042586.10 ENSMUST00000218605.2 |
Tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr17_+_46421908 | 0.38 |
ENSMUST00000024763.10
ENSMUST00000123646.2 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr11_-_119190830 | 0.38 |
ENSMUST00000106253.2
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr17_+_28059129 | 0.37 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr3_-_65300000 | 0.37 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr4_-_116664729 | 0.36 |
ENSMUST00000106455.8
ENSMUST00000030451.10 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr6_-_120799672 | 0.36 |
ENSMUST00000019354.11
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr14_-_30723292 | 0.35 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr4_+_118266526 | 0.35 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr10_+_80008768 | 0.35 |
ENSMUST00000041882.7
|
Fam174c
|
family with sequence similarity 174, member C |
| chr8_+_12965876 | 0.35 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr14_-_101877870 | 0.35 |
ENSMUST00000168587.3
|
Commd6
|
COMM domain containing 6 |
| chr17_+_26882171 | 0.35 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr7_+_81412695 | 0.34 |
ENSMUST00000133034.2
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr11_-_120515799 | 0.34 |
ENSMUST00000106183.3
ENSMUST00000080202.12 |
Sirt7
|
sirtuin 7 |
| chr9_-_22028370 | 0.33 |
ENSMUST00000213233.2
|
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr13_-_95511837 | 0.33 |
ENSMUST00000022189.9
|
Aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr5_-_110928436 | 0.33 |
ENSMUST00000149208.2
ENSMUST00000031483.15 ENSMUST00000086643.12 ENSMUST00000170468.8 ENSMUST00000031481.13 |
Pus1
|
pseudouridine synthase 1 |
| chr4_+_118266582 | 0.33 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr9_-_21061196 | 0.32 |
ENSMUST00000215296.2
ENSMUST00000019615.11 |
Cdc37
|
cell division cycle 37 |
| chr7_-_105460172 | 0.32 |
ENSMUST00000033170.7
ENSMUST00000176498.2 ENSMUST00000176887.2 ENSMUST00000124482.3 |
Mrpl17
|
mitochondrial ribosomal protein L17 |
| chr11_+_106146966 | 0.32 |
ENSMUST00000021049.9
|
Psmc5
|
protease (prosome, macropain) 26S subunit, ATPase 5 |
| chr19_+_6135013 | 0.32 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr18_+_34994253 | 0.32 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr5_-_139812592 | 0.31 |
ENSMUST00000182602.2
ENSMUST00000031531.14 |
Psmg3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr3_+_157239988 | 0.31 |
ENSMUST00000029831.16
ENSMUST00000106057.8 |
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr12_-_55126882 | 0.31 |
ENSMUST00000021406.6
|
2700097O09Rik
|
RIKEN cDNA 2700097O09 gene |
| chr19_+_8849000 | 0.31 |
ENSMUST00000096255.7
|
Ubxn1
|
UBX domain protein 1 |
| chr6_-_120799500 | 0.31 |
ENSMUST00000204699.2
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr11_-_119190896 | 0.31 |
ENSMUST00000026667.15
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr2_-_93841062 | 0.30 |
ENSMUST00000040005.13
ENSMUST00000126378.8 |
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr7_+_81412673 | 0.30 |
ENSMUST00000042166.11
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr9_+_107440445 | 0.30 |
ENSMUST00000010198.5
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr12_+_55276953 | 0.29 |
ENSMUST00000218879.2
|
Srp54c
|
signal recognition particle 54C |
| chr17_+_28059099 | 0.29 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr15_+_58805605 | 0.29 |
ENSMUST00000022980.5
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr10_+_128641663 | 0.28 |
ENSMUST00000218511.2
|
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr3_-_65299967 | 0.28 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr8_-_22888604 | 0.28 |
ENSMUST00000033871.8
|
Slc25a15
|
solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 |
| chr9_+_107440484 | 0.28 |
ENSMUST00000193418.2
|
Tusc2
|
tumor suppressor 2, mitochondrial calcium regulator |
| chr19_-_44994824 | 0.28 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr7_-_45084012 | 0.28 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr3_+_103875261 | 0.27 |
ENSMUST00000117150.8
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr7_+_126832399 | 0.27 |
ENSMUST00000056232.7
|
Zfp553
|
zinc finger protein 553 |
| chr4_+_45297127 | 0.27 |
ENSMUST00000044673.9
ENSMUST00000144781.3 |
Trmt10b
|
tRNA methyltransferase 10B |
| chr10_-_67120959 | 0.27 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chrX_+_7750483 | 0.27 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr10_-_30476658 | 0.27 |
ENSMUST00000019927.7
|
Trmt11
|
tRNA methyltransferase 11 |
| chr4_-_98271469 | 0.26 |
ENSMUST00000143116.2
ENSMUST00000030292.12 ENSMUST00000102793.11 |
Tm2d1
|
TM2 domain containing 1 |
| chr2_+_29825427 | 0.26 |
ENSMUST00000019859.9
|
Gle1
|
GLE1 RNA export mediator (yeast) |
| chr10_+_128641520 | 0.26 |
ENSMUST00000219508.2
ENSMUST00000026410.2 |
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr14_-_101878106 | 0.26 |
ENSMUST00000100339.9
|
Commd6
|
COMM domain containing 6 |
| chr10_-_31485180 | 0.26 |
ENSMUST00000081989.8
|
Rnf217
|
ring finger protein 217 |
| chr11_-_20062876 | 0.26 |
ENSMUST00000000137.8
|
Actr2
|
ARP2 actin-related protein 2 |
| chrX_+_7750558 | 0.26 |
ENSMUST00000208640.2
ENSMUST00000207114.2 ENSMUST00000208633.2 ENSMUST00000208397.2 ENSMUST00000153620.3 ENSMUST00000123277.8 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr7_+_126832215 | 0.25 |
ENSMUST00000106312.4
|
Zfp553
|
zinc finger protein 553 |
| chr4_+_116664911 | 0.25 |
ENSMUST00000102699.8
ENSMUST00000130359.8 |
Mutyh
|
mutY DNA glycosylase |
| chr19_-_5416339 | 0.25 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr17_+_47022384 | 0.25 |
ENSMUST00000002840.9
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr17_-_26063391 | 0.25 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr5_+_24791719 | 0.25 |
ENSMUST00000088295.9
ENSMUST00000121863.5 |
Chpf2
|
chondroitin polymerizing factor 2 |
| chr11_+_88095222 | 0.24 |
ENSMUST00000118784.8
ENSMUST00000139170.8 ENSMUST00000107915.10 ENSMUST00000144070.3 |
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr15_+_79555272 | 0.24 |
ENSMUST00000127292.2
|
Tomm22
|
translocase of outer mitochondrial membrane 22 |
| chr10_-_14581203 | 0.24 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr2_-_30857858 | 0.24 |
ENSMUST00000028200.9
|
Tor1a
|
torsin family 1, member A (torsin A) |
| chrX_+_7750261 | 0.23 |
ENSMUST00000115660.12
|
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr11_-_120348762 | 0.23 |
ENSMUST00000137632.2
ENSMUST00000044007.3 |
Oxld1
|
oxidoreductase like domain containing 1 |
| chr11_+_120839288 | 0.23 |
ENSMUST00000070653.13
|
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr15_+_76215488 | 0.23 |
ENSMUST00000172281.8
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr5_-_110417682 | 0.23 |
ENSMUST00000196381.2
ENSMUST00000059229.16 ENSMUST00000112505.9 |
Pgam5
|
phosphoglycerate mutase family member 5 |
| chr8_+_107783502 | 0.23 |
ENSMUST00000034392.13
ENSMUST00000170962.3 |
Nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr11_+_87628356 | 0.23 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr10_-_80156337 | 0.23 |
ENSMUST00000020341.9
|
2310011J03Rik
|
RIKEN cDNA 2310011J03 gene |
| chr2_+_48704121 | 0.23 |
ENSMUST00000063886.4
|
Acvr2a
|
activin receptor IIA |
| chr4_-_124744943 | 0.23 |
ENSMUST00000185036.2
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr13_-_4329421 | 0.23 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr11_+_88095206 | 0.22 |
ENSMUST00000024486.14
|
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr15_-_85695810 | 0.22 |
ENSMUST00000144067.8
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr8_-_108315024 | 0.22 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr3_+_89337568 | 0.22 |
ENSMUST00000060061.11
|
Pygo2
|
pygopus 2 |
| chr19_+_8848924 | 0.22 |
ENSMUST00000238036.2
|
Ubxn1
|
UBX domain protein 1 |
| chr9_+_99457829 | 0.21 |
ENSMUST00000066650.12
ENSMUST00000148987.8 |
Dbr1
|
debranching RNA lariats 1 |
| chr1_+_58625539 | 0.21 |
ENSMUST00000027193.9
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr16_+_64672334 | 0.21 |
ENSMUST00000067744.8
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr12_+_104998895 | 0.21 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr8_+_110944575 | 0.21 |
ENSMUST00000056972.6
|
Cmtr2
|
cap methyltransferase 2 |
| chr12_+_102724223 | 0.21 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr15_+_5215000 | 0.21 |
ENSMUST00000118193.8
ENSMUST00000022751.15 |
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr12_+_78908466 | 0.21 |
ENSMUST00000071230.8
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr11_+_61544085 | 0.21 |
ENSMUST00000004959.3
|
Grap
|
GRB2-related adaptor protein |
| chr11_+_119989736 | 0.21 |
ENSMUST00000106223.4
ENSMUST00000185558.2 |
Ndufaf8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr12_-_87435091 | 0.21 |
ENSMUST00000021424.5
|
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr15_+_76215431 | 0.20 |
ENSMUST00000023221.13
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr4_+_156028281 | 0.20 |
ENSMUST00000103175.8
ENSMUST00000166489.8 ENSMUST00000024056.10 ENSMUST00000136492.2 ENSMUST00000105583.9 ENSMUST00000152536.8 ENSMUST00000118192.8 |
Ube2j2
|
ubiquitin-conjugating enzyme E2J 2 |
| chr4_-_59438633 | 0.20 |
ENSMUST00000040166.14
ENSMUST00000107544.2 |
Susd1
|
sushi domain containing 1 |
| chr6_-_115785695 | 0.20 |
ENSMUST00000081840.6
|
Rpl32
|
ribosomal protein L32 |
| chr2_-_145776934 | 0.20 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr4_+_124960465 | 0.20 |
ENSMUST00000052183.7
|
Snip1
|
Smad nuclear interacting protein 1 |
| chr19_+_3332901 | 0.20 |
ENSMUST00000025745.10
ENSMUST00000025743.7 |
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr4_-_133005039 | 0.20 |
ENSMUST00000105907.9
|
Tmem222
|
transmembrane protein 222 |
| chr12_+_55201889 | 0.20 |
ENSMUST00000110708.4
|
Srp54b
|
signal recognition particle 54B |
| chr1_+_75498162 | 0.20 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr4_+_107035615 | 0.20 |
ENSMUST00000128123.3
ENSMUST00000106753.3 |
Tmem59
|
transmembrane protein 59 |
| chr19_-_27407206 | 0.20 |
ENSMUST00000076219.6
|
Pum3
|
pumilio RNA-binding family member 3 |
| chr11_+_96932379 | 0.20 |
ENSMUST00000001485.10
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr11_-_79853200 | 0.20 |
ENSMUST00000108241.8
ENSMUST00000043152.6 |
Utp6
|
UTP6 small subunit processome component |
| chrX_+_99669343 | 0.19 |
ENSMUST00000048962.4
|
Kif4
|
kinesin family member 4 |
| chr3_+_157239740 | 0.19 |
ENSMUST00000106058.8
|
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr17_-_50497682 | 0.19 |
ENSMUST00000044503.14
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr19_+_8848876 | 0.19 |
ENSMUST00000166407.9
|
Ubxn1
|
UBX domain protein 1 |
| chr2_-_132657891 | 0.19 |
ENSMUST00000039554.7
|
Trmt6
|
tRNA methyltransferase 6 |
| chr5_-_149559636 | 0.19 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr12_+_55126999 | 0.18 |
ENSMUST00000220578.2
ENSMUST00000221655.2 |
Srp54a
|
signal recognition particle 54A |
| chr19_+_5416769 | 0.18 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr9_-_22028419 | 0.18 |
ENSMUST00000214394.2
ENSMUST00000013966.8 |
Elof1
|
ELF1 homolog, elongation factor 1 |
| chr8_-_78337297 | 0.18 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr15_-_34443355 | 0.18 |
ENSMUST00000009039.6
|
Rpl30
|
ribosomal protein L30 |
| chr14_+_55909692 | 0.18 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr18_+_31922173 | 0.18 |
ENSMUST00000025106.5
ENSMUST00000234146.2 |
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr4_+_118477994 | 0.18 |
ENSMUST00000030501.15
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr6_-_88495835 | 0.18 |
ENSMUST00000032168.7
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr4_-_48279543 | 0.17 |
ENSMUST00000030028.5
|
Erp44
|
endoplasmic reticulum protein 44 |
| chr19_+_44994905 | 0.17 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr5_-_149559667 | 0.17 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr10_+_128641398 | 0.17 |
ENSMUST00000217745.2
|
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr11_-_115590318 | 0.17 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr11_-_84710276 | 0.17 |
ENSMUST00000018549.8
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr8_-_84059048 | 0.17 |
ENSMUST00000177594.8
ENSMUST00000053902.4 |
Elmod2
|
ELMO/CED-12 domain containing 2 |
| chr1_-_125363272 | 0.16 |
ENSMUST00000178474.8
|
Actr3
|
ARP3 actin-related protein 3 |
| chr7_+_65693350 | 0.16 |
ENSMUST00000206065.2
|
Gm45213
|
predicted gene 45213 |
| chr9_-_57552392 | 0.16 |
ENSMUST00000216934.2
|
Csk
|
c-src tyrosine kinase |
| chr7_+_79910948 | 0.16 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr7_-_105393519 | 0.16 |
ENSMUST00000141116.2
|
Taf10
|
TATA-box binding protein associated factor 10 |
| chr2_-_93841152 | 0.16 |
ENSMUST00000111240.8
|
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr14_+_55829165 | 0.16 |
ENSMUST00000019443.15
|
Rnf31
|
ring finger protein 31 |
| chr5_+_147797258 | 0.16 |
ENSMUST00000031654.10
|
Pomp
|
proteasome maturation protein |
| chr4_-_119177614 | 0.16 |
ENSMUST00000147077.8
ENSMUST00000056458.14 ENSMUST00000106321.9 ENSMUST00000106319.8 ENSMUST00000106317.2 ENSMUST00000106318.8 |
Ppih
|
peptidyl prolyl isomerase H |
| chrX_-_93256244 | 0.16 |
ENSMUST00000113922.2
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr1_-_36283326 | 0.15 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr1_-_58625431 | 0.15 |
ENSMUST00000161000.2
ENSMUST00000161600.8 |
Fam126b
|
family with sequence similarity 126, member B |
| chr11_+_96932395 | 0.15 |
ENSMUST00000054252.5
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr11_-_16458069 | 0.15 |
ENSMUST00000109641.2
|
Sec61g
|
SEC61, gamma subunit |
| chr1_-_179373826 | 0.15 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr11_-_106146868 | 0.15 |
ENSMUST00000021048.7
|
Ftsj3
|
FtsJ RNA methyltransferase homolog 3 (E. coli) |
| chr3_-_5641171 | 0.15 |
ENSMUST00000071280.8
ENSMUST00000195855.6 ENSMUST00000165309.8 ENSMUST00000164828.8 |
Pex2
|
peroxisomal biogenesis factor 2 |
| chr1_+_167135933 | 0.14 |
ENSMUST00000195015.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr15_+_5215180 | 0.14 |
ENSMUST00000081640.12
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr11_+_94544593 | 0.14 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr1_+_4878460 | 0.14 |
ENSMUST00000131119.2
|
Lypla1
|
lysophospholipase 1 |
| chr2_+_152068729 | 0.14 |
ENSMUST00000099224.10
ENSMUST00000124791.8 ENSMUST00000133119.2 |
Csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr9_+_40103598 | 0.14 |
ENSMUST00000026693.14
ENSMUST00000168832.2 |
Zfp202
|
zinc finger protein 202 |
| chr11_+_50101717 | 0.13 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr18_+_12261798 | 0.13 |
ENSMUST00000025270.8
|
Riok3
|
RIO kinase 3 |
| chr5_+_124690908 | 0.13 |
ENSMUST00000071057.14
ENSMUST00000111438.2 |
Ddx55
|
DEAD box helicase 55 |
| chr2_+_132528851 | 0.13 |
ENSMUST00000061891.11
|
Shld1
|
shieldin complex subunit 1 |
| chr4_-_41314877 | 0.13 |
ENSMUST00000030145.9
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chrX_-_99669507 | 0.13 |
ENSMUST00000059099.7
|
Pdzd11
|
PDZ domain containing 11 |
| chr11_+_5905693 | 0.13 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr19_+_44551280 | 0.13 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr19_+_4204605 | 0.13 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr8_-_111573401 | 0.12 |
ENSMUST00000042012.7
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr15_-_81931783 | 0.12 |
ENSMUST00000080622.9
|
Snu13
|
SNU13 homolog, small nuclear ribonucleoprotein (U4/U6.U5) |
| chr6_+_124785621 | 0.12 |
ENSMUST00000047760.10
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr11_-_115590133 | 0.12 |
ENSMUST00000106499.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr1_+_4878046 | 0.12 |
ENSMUST00000027036.11
ENSMUST00000150971.8 ENSMUST00000119612.9 ENSMUST00000137887.8 ENSMUST00000115529.8 |
Lypla1
|
lysophospholipase 1 |
| chr2_+_127050281 | 0.12 |
ENSMUST00000103220.4
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr4_-_136613498 | 0.12 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr7_-_80597479 | 0.12 |
ENSMUST00000026818.12
ENSMUST00000117383.8 ENSMUST00000119980.2 |
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr2_+_32425327 | 0.12 |
ENSMUST00000133512.2
ENSMUST00000048375.6 |
Fam102a
|
family with sequence similarity 102, member A |
| chr4_-_46389391 | 0.11 |
ENSMUST00000086563.11
ENSMUST00000030015.6 |
Trmo
|
tRNA methyltransferase O |
| chr4_-_45320579 | 0.11 |
ENSMUST00000030003.10
|
Exosc3
|
exosome component 3 |
| chr2_-_75808508 | 0.11 |
ENSMUST00000099995.5
|
Ttc30a2
|
tetratricopeptide repeat domain 30A2 |
| chr13_+_20274708 | 0.11 |
ENSMUST00000072519.7
|
Elmo1
|
engulfment and cell motility 1 |
| chr8_+_105066980 | 0.11 |
ENSMUST00000211885.2
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr4_-_40948196 | 0.11 |
ENSMUST00000030125.5
ENSMUST00000108089.8 ENSMUST00000191273.7 |
Bag1
|
BCL2-associated athanogene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Etv6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0052572 | interleukin-15 production(GO:0032618) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.2 | 0.7 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.7 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.1 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.7 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.3 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.5 | GO:0030576 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.6 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.3 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.5 | GO:0035553 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.5 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.5 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 1.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 0.2 | GO:0046038 | GMP catabolic process(GO:0046038) |
| 0.1 | 0.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.2 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 1.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0051940 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.1 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.3 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.1 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.8 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.9 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 0.7 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 0.7 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.2 | 0.5 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 1.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.3 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 1.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.4 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.7 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |