Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Figla
Z-value: 0.91
Transcription factors associated with Figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
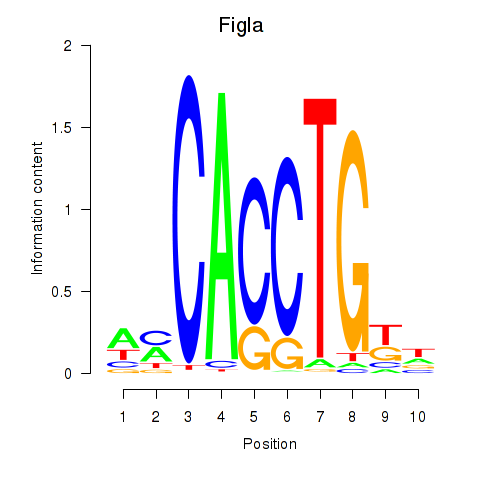
Figla
|
ENSMUSG00000030001.5 | folliculogenesis specific basic helix-loop-helix |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Figla | mm39_v1_chr6_+_85994173_85994214 | -0.28 | 6.5e-01 | Click! |
Activity profile of Figla motif
Sorted Z-values of Figla motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_122441719 | 0.71 |
ENSMUST00000028624.9
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_-_126866682 | 0.66 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr11_-_48836975 | 0.66 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr11_+_101207743 | 0.62 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr7_-_144493560 | 0.56 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr5_-_138169509 | 0.52 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr9_-_108443916 | 0.52 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr11_+_53661251 | 0.50 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr8_+_120121612 | 0.47 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr3_+_51324022 | 0.44 |
ENSMUST00000192419.6
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr9_-_44253630 | 0.43 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr9_-_108444561 | 0.42 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr2_-_32150151 | 0.41 |
ENSMUST00000002625.15
|
Uck1
|
uridine-cytidine kinase 1 |
| chr5_-_138170077 | 0.40 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_+_123214332 | 0.39 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr15_+_88703786 | 0.38 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr3_-_89905547 | 0.37 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr4_-_152122891 | 0.36 |
ENSMUST00000030792.2
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chr16_+_49620883 | 0.36 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr14_-_55114989 | 0.35 |
ENSMUST00000168622.2
ENSMUST00000177403.2 |
Ppp1r3e
|
protein phosphatase 1, regulatory subunit 3E |
| chr14_-_31552335 | 0.34 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr11_-_97171258 | 0.34 |
ENSMUST00000165216.8
|
Npepps
|
aminopeptidase puromycin sensitive |
| chr13_-_100922910 | 0.32 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr10_+_75768964 | 0.32 |
ENSMUST00000219839.2
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr3_+_106393348 | 0.32 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr15_-_78290038 | 0.31 |
ENSMUST00000058659.9
|
Tst
|
thiosulfate sulfurtransferase, mitochondrial |
| chr19_+_5088854 | 0.31 |
ENSMUST00000053705.8
ENSMUST00000235776.2 |
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chr14_+_105496008 | 0.30 |
ENSMUST00000181969.8
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr6_-_40413056 | 0.30 |
ENSMUST00000039008.10
ENSMUST00000101492.10 |
Dennd11
|
DENN domain containing 11 |
| chr4_+_106418224 | 0.30 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr19_+_36325683 | 0.29 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr10_-_126866658 | 0.29 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr8_-_48443525 | 0.29 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr5_+_122347912 | 0.29 |
ENSMUST00000143560.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr3_+_95496270 | 0.28 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr10_+_26105605 | 0.28 |
ENSMUST00000218301.2
ENSMUST00000164660.8 ENSMUST00000060716.6 |
Samd3
|
sterile alpha motif domain containing 3 |
| chr15_-_81614063 | 0.27 |
ENSMUST00000171115.3
ENSMUST00000170134.9 ENSMUST00000052374.13 |
Rangap1
|
RAN GTPase activating protein 1 |
| chr1_-_52271455 | 0.27 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr14_+_105496147 | 0.26 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr15_+_102381705 | 0.26 |
ENSMUST00000229958.2
ENSMUST00000229184.2 ENSMUST00000230728.2 ENSMUST00000230114.2 |
Pcbp2
|
poly(rC) binding protein 2 |
| chr8_-_71834543 | 0.26 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr9_+_107852733 | 0.26 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr17_+_35413415 | 0.26 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr11_+_99748741 | 0.25 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr3_+_94882142 | 0.24 |
ENSMUST00000167008.8
ENSMUST00000107251.9 |
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr7_-_126817475 | 0.24 |
ENSMUST00000106313.8
ENSMUST00000142356.3 |
Septin1
|
septin 1 |
| chr16_+_20430335 | 0.24 |
ENSMUST00000115522.10
ENSMUST00000119224.8 ENSMUST00000120394.8 ENSMUST00000079600.12 |
Eef1akmt4
Gm49333
|
EEF1A lysine methyltransferase 4 predicted gene, 49333 |
| chr11_+_97692738 | 0.24 |
ENSMUST00000127033.9
|
Lasp1
|
LIM and SH3 protein 1 |
| chr8_-_70355208 | 0.23 |
ENSMUST00000110167.5
|
Ndufa13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| chr9_+_107445101 | 0.23 |
ENSMUST00000192887.6
ENSMUST00000195752.6 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr12_-_40087393 | 0.22 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr5_+_138170259 | 0.22 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr3_+_65435825 | 0.22 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr11_+_87628356 | 0.22 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr11_-_82761954 | 0.22 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr3_+_88523730 | 0.22 |
ENSMUST00000175779.8
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr3_-_89152320 | 0.21 |
ENSMUST00000107464.8
ENSMUST00000090924.13 |
Trim46
|
tripartite motif-containing 46 |
| chr2_+_122461079 | 0.21 |
ENSMUST00000239506.1
|
SPATA5L1
|
spermatosis associated 5-like 1 |
| chr11_+_120675324 | 0.21 |
ENSMUST00000172809.2
|
Gps1
|
G protein pathway suppressor 1 |
| chr5_-_140816111 | 0.20 |
ENSMUST00000000153.9
|
Gna12
|
guanine nucleotide binding protein, alpha 12 |
| chr17_-_29566774 | 0.20 |
ENSMUST00000095427.12
ENSMUST00000118366.9 |
Mtch1
|
mitochondrial carrier 1 |
| chr13_+_21364330 | 0.20 |
ENSMUST00000223065.2
|
Trim27
|
tripartite motif-containing 27 |
| chr3_+_94882038 | 0.20 |
ENSMUST00000072287.12
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr9_+_57468217 | 0.20 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr11_-_97171294 | 0.18 |
ENSMUST00000167806.8
ENSMUST00000172108.8 ENSMUST00000001480.14 |
Npepps
|
aminopeptidase puromycin sensitive |
| chr11_+_3282424 | 0.18 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_-_52225763 | 0.18 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr5_+_123887759 | 0.18 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr3_-_107603778 | 0.18 |
ENSMUST00000029490.15
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1 |
| chr1_+_156386414 | 0.18 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr11_+_120675131 | 0.18 |
ENSMUST00000116305.8
|
Gps1
|
G protein pathway suppressor 1 |
| chr1_-_59158902 | 0.18 |
ENSMUST00000087475.11
|
Tmem237
|
transmembrane protein 237 |
| chr19_-_12478803 | 0.17 |
ENSMUST00000045521.9
|
Dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr11_-_77404160 | 0.17 |
ENSMUST00000060417.11
|
Trp53i13
|
transformation related protein 53 inducible protein 13 |
| chr3_+_95496239 | 0.17 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr19_+_56536685 | 0.17 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr12_-_40088024 | 0.17 |
ENSMUST00000101472.4
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr15_-_89007290 | 0.17 |
ENSMUST00000109353.9
|
Tubgcp6
|
tubulin, gamma complex associated protein 6 |
| chr11_+_115714853 | 0.16 |
ENSMUST00000103032.11
ENSMUST00000133250.8 ENSMUST00000177736.8 |
Llgl2
|
LLGL2 scribble cell polarity complex component |
| chr17_+_34258411 | 0.16 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr7_-_24771717 | 0.16 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr15_-_78657640 | 0.16 |
ENSMUST00000018313.6
|
Mfng
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_+_34818709 | 0.16 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr10_-_21036792 | 0.16 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr1_-_182929025 | 0.15 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr9_-_44253588 | 0.15 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr15_+_99615396 | 0.15 |
ENSMUST00000023760.13
ENSMUST00000162194.2 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr17_+_24692858 | 0.14 |
ENSMUST00000054946.10
ENSMUST00000238986.2 ENSMUST00000164508.2 |
Bricd5
|
BRICHOS domain containing 5 |
| chr9_-_107960528 | 0.14 |
ENSMUST00000159372.3
ENSMUST00000160249.8 |
Rnf123
|
ring finger protein 123 |
| chr4_-_135221926 | 0.14 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr8_+_108162985 | 0.14 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr6_+_39358036 | 0.14 |
ENSMUST00000031986.5
|
Rab19
|
RAB19, member RAS oncogene family |
| chr4_-_133600308 | 0.14 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr12_-_34578842 | 0.14 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr2_-_34716083 | 0.13 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr4_-_133599616 | 0.13 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr7_+_130179063 | 0.13 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr9_-_20809888 | 0.13 |
ENSMUST00000004206.10
|
Eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr5_+_122348140 | 0.13 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr4_-_42168603 | 0.13 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr4_-_42665763 | 0.13 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_-_97171185 | 0.13 |
ENSMUST00000168743.8
|
Npepps
|
aminopeptidase puromycin sensitive |
| chr13_+_76532470 | 0.12 |
ENSMUST00000125209.8
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr7_+_27259895 | 0.12 |
ENSMUST00000187032.2
|
2310022A10Rik
|
RIKEN cDNA 2310022A10 gene |
| chr5_+_137627431 | 0.12 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr1_-_128030148 | 0.12 |
ENSMUST00000086614.12
|
Zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr16_-_43709968 | 0.12 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chrX_-_73325204 | 0.11 |
ENSMUST00000114189.10
ENSMUST00000119361.4 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr17_-_66175046 | 0.11 |
ENSMUST00000233399.2
|
Ralbp1
|
ralA binding protein 1 |
| chr7_-_126817639 | 0.11 |
ENSMUST00000152267.8
ENSMUST00000106314.8 |
Septin1
|
septin 1 |
| chr11_-_99549786 | 0.11 |
ENSMUST00000092695.5
|
Gm11563
|
predicted gene 11563 |
| chrX_-_74460168 | 0.11 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr16_+_16031182 | 0.11 |
ENSMUST00000039408.3
|
Pkp2
|
plakophilin 2 |
| chrX_-_36253309 | 0.11 |
ENSMUST00000060474.14
ENSMUST00000053456.11 ENSMUST00000115239.10 |
Septin6
|
septin 6 |
| chrX_-_73412078 | 0.10 |
ENSMUST00000155676.8
|
Ubl4a
|
ubiquitin-like 4A |
| chr3_-_89905927 | 0.10 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr8_-_86159398 | 0.10 |
ENSMUST00000047749.7
|
4921524J17Rik
|
RIKEN cDNA 4921524J17 gene |
| chr9_+_63509925 | 0.10 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr18_-_73948501 | 0.10 |
ENSMUST00000025439.5
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr5_-_138169476 | 0.10 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr16_-_52272828 | 0.10 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr11_-_99556781 | 0.10 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr13_-_114524586 | 0.10 |
ENSMUST00000232101.2
ENSMUST00000225978.3 |
Ndufs4
|
NADH:ubiquinone oxidoreductase core subunit S4 |
| chr2_+_167922924 | 0.10 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr17_-_88105422 | 0.10 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr11_-_69576363 | 0.09 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_-_126362372 | 0.09 |
ENSMUST00000097888.10
ENSMUST00000239399.2 |
Ago1
|
argonaute RISC catalytic subunit 1 |
| chr5_-_73349191 | 0.09 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr19_-_56536646 | 0.09 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr2_+_164611812 | 0.09 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr11_+_99839246 | 0.09 |
ENSMUST00000105052.2
|
Krtap9-5
|
keratin associated protein 9-5 |
| chr1_+_59555423 | 0.09 |
ENSMUST00000114243.8
|
Gm973
|
predicted gene 973 |
| chr15_-_97629209 | 0.08 |
ENSMUST00000100249.10
|
Endou
|
endonuclease, polyU-specific |
| chr10_-_21036824 | 0.08 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr15_+_41652777 | 0.08 |
ENSMUST00000230778.2
ENSMUST00000022918.15 ENSMUST00000090095.13 |
Oxr1
|
oxidation resistance 1 |
| chr8_-_47805383 | 0.08 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr3_+_84832783 | 0.08 |
ENSMUST00000107675.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr2_-_52225146 | 0.08 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr1_+_128069716 | 0.08 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr1_-_133352115 | 0.08 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr5_+_150679760 | 0.07 |
ENSMUST00000110486.2
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr4_-_116228921 | 0.07 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr15_-_63869818 | 0.07 |
ENSMUST00000164532.3
|
Cyrib
|
CYFIP related Rac1 interactor B |
| chr11_+_19874403 | 0.07 |
ENSMUST00000093298.12
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr15_+_98532624 | 0.07 |
ENSMUST00000003442.9
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr9_+_44966464 | 0.07 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr5_+_146168020 | 0.07 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr4_+_101944740 | 0.07 |
ENSMUST00000106911.8
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_-_9029097 | 0.07 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr9_+_105535585 | 0.07 |
ENSMUST00000186943.2
|
Pik3r4
|
phosphoinositide-3-kinase regulatory subunit 4 |
| chr7_-_24919247 | 0.07 |
ENSMUST00000058702.7
|
Dedd2
|
death effector domain-containing DNA binding protein 2 |
| chr9_+_102597486 | 0.07 |
ENSMUST00000130602.2
|
Amotl2
|
angiomotin-like 2 |
| chr5_+_20112500 | 0.06 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_-_36011378 | 0.06 |
ENSMUST00000119825.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr2_-_62313981 | 0.06 |
ENSMUST00000136686.2
ENSMUST00000102733.10 |
Gcg
|
glucagon |
| chr2_-_91025492 | 0.06 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr3_-_116601700 | 0.06 |
ENSMUST00000159742.8
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr17_-_32607859 | 0.06 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr3_+_65435878 | 0.06 |
ENSMUST00000130705.2
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr2_+_20742115 | 0.06 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr11_+_109541400 | 0.06 |
ENSMUST00000106676.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr6_-_70313491 | 0.06 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr11_+_99770013 | 0.06 |
ENSMUST00000078442.4
|
Gm11567
|
predicted gene 11567 |
| chr11_-_20781009 | 0.06 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr17_-_24869241 | 0.05 |
ENSMUST00000234557.2
ENSMUST00000002572.6 |
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr9_+_43978290 | 0.05 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr6_-_8778439 | 0.05 |
ENSMUST00000115520.8
ENSMUST00000038403.12 ENSMUST00000115518.8 |
Ica1
|
islet cell autoantigen 1 |
| chr11_-_99689758 | 0.05 |
ENSMUST00000105057.2
|
Gm11569
|
predicted gene 11569 |
| chr19_-_11243530 | 0.05 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_110956980 | 0.05 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr8_+_96442509 | 0.05 |
ENSMUST00000034096.6
|
Setd6
|
SET domain containing 6 |
| chrX_-_74460137 | 0.05 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr17_-_32639936 | 0.05 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr7_-_12096691 | 0.05 |
ENSMUST00000086228.3
|
Vmn1r84
|
vomeronasal 1 receptor 84 |
| chr9_-_50657800 | 0.04 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr16_-_95260104 | 0.04 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr7_+_45289391 | 0.04 |
ENSMUST00000148532.4
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr11_-_69496655 | 0.04 |
ENSMUST00000047889.13
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr17_-_34247016 | 0.04 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr11_+_99676017 | 0.04 |
ENSMUST00000105059.4
|
Krtap4-9
|
keratin associated protein 4-9 |
| chr1_+_152626273 | 0.04 |
ENSMUST00000068875.5
|
Apobec4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr2_-_76812799 | 0.04 |
ENSMUST00000011934.13
ENSMUST00000099981.10 ENSMUST00000099980.10 ENSMUST00000111882.9 ENSMUST00000140091.8 |
Ttn
|
titin |
| chr2_-_170248421 | 0.04 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr2_+_122479770 | 0.04 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr10_-_75768302 | 0.04 |
ENSMUST00000120281.8
ENSMUST00000000924.13 |
Mmp11
|
matrix metallopeptidase 11 |
| chr14_-_104705420 | 0.03 |
ENSMUST00000053016.10
|
Pou4f1
|
POU domain, class 4, transcription factor 1 |
| chr19_-_56378459 | 0.03 |
ENSMUST00000040711.15
ENSMUST00000095947.11 ENSMUST00000073536.13 |
Nrap
|
nebulin-related anchoring protein |
| chr11_-_99700718 | 0.03 |
ENSMUST00000050106.5
|
Krtap4-13
|
keratin associated protein 4-13 |
| chr11_-_99494134 | 0.03 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr6_-_8778106 | 0.03 |
ENSMUST00000151758.2
ENSMUST00000115519.8 ENSMUST00000153390.8 |
Ica1
|
islet cell autoantigen 1 |
| chr6_-_3494587 | 0.03 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr19_-_56378309 | 0.03 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr11_-_99519057 | 0.03 |
ENSMUST00000081007.7
|
Krtap4-1
|
keratin associated protein 4-1 |
| chr11_+_99805555 | 0.03 |
ENSMUST00000105053.2
|
Gm11565
|
predicted gene 11565 |
| chr7_+_126811831 | 0.03 |
ENSMUST00000127710.3
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr6_-_70116066 | 0.03 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr11_-_99684173 | 0.03 |
ENSMUST00000105058.2
|
Gm11596
|
predicted gene 11596 |
| chr14_+_65504067 | 0.03 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr13_+_113346193 | 0.03 |
ENSMUST00000038144.9
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr2_+_26969384 | 0.03 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Figla
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.7 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 0.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.3 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.5 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.3 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 | 0.6 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 0.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.0 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0034334 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.0 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.4 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |