Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Fli1
Z-value: 1.02
Transcription factors associated with Fli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fli1
|
ENSMUSG00000016087.14 | Friend leukemia integration 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fli1 | mm39_v1_chr9_-_32454157_32454305 | 0.99 | 1.7e-03 | Click! |
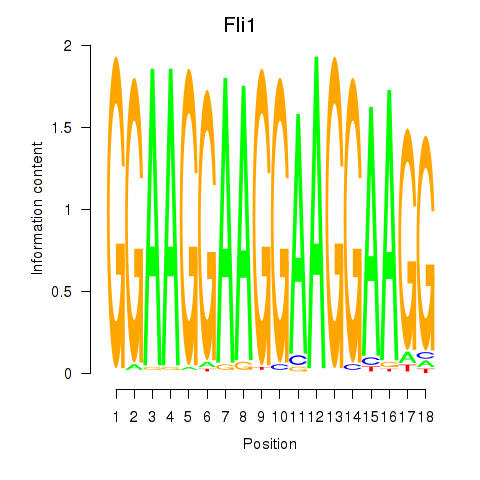
Activity profile of Fli1 motif
Sorted Z-values of Fli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_104506216 | 3.09 |
ENSMUST00000067695.8
|
Usp17la
|
ubiquitin specific peptidase 17-like A |
| chrX_-_135769285 | 2.37 |
ENSMUST00000058814.7
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr7_-_15978077 | 1.71 |
ENSMUST00000238893.2
ENSMUST00000171425.4 |
C5ar2
|
complement component 5a receptor 2 |
| chr7_-_6525801 | 1.63 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr2_-_120867529 | 1.26 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr16_+_3648742 | 1.20 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr7_+_11608557 | 0.71 |
ENSMUST00000227611.2
ENSMUST00000226622.2 ENSMUST00000228646.2 ENSMUST00000226855.2 ENSMUST00000228268.2 ENSMUST00000228463.2 |
Vmn1r75
|
vomeronasal 1 receptor 75 |
| chr6_+_57183497 | 0.70 |
ENSMUST00000227298.2
|
Vmn1r13
|
vomeronasal 1 receptor 13 |
| chr17_+_29077385 | 0.69 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr14_-_7666348 | 0.66 |
ENSMUST00000224491.2
|
Nek10
|
NIMA (never in mitosis gene a)- related kinase 10 |
| chr8_+_23525101 | 0.63 |
ENSMUST00000117662.8
ENSMUST00000117296.8 ENSMUST00000141784.9 |
Ank1
|
ankyrin 1, erythroid |
| chr6_-_28831746 | 0.56 |
ENSMUST00000062304.7
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr11_+_16702203 | 0.48 |
ENSMUST00000102884.10
ENSMUST00000020329.13 |
Egfr
|
epidermal growth factor receptor |
| chr1_+_174000304 | 0.45 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr9_-_108991088 | 0.44 |
ENSMUST00000199540.2
ENSMUST00000198076.5 ENSMUST00000054925.13 |
Fbxw21
|
F-box and WD-40 domain protein 21 |
| chr7_-_19410749 | 0.42 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr17_-_53846438 | 0.40 |
ENSMUST00000056198.4
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr2_-_38177359 | 0.40 |
ENSMUST00000102787.10
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr10_-_71121083 | 0.39 |
ENSMUST00000020085.7
|
Ube2d1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr10_-_62561865 | 0.39 |
ENSMUST00000133371.8
|
Stox1
|
storkhead box 1 |
| chr7_+_11905206 | 0.38 |
ENSMUST00000226953.2
ENSMUST00000227530.2 |
Vmn1r79
|
vomeronasal 1 receptor 79 |
| chr7_+_103064279 | 0.38 |
ENSMUST00000106892.2
|
Usp17lc
|
ubiquitin specific peptidase 17-like C |
| chr13_+_99321241 | 0.37 |
ENSMUST00000056558.11
|
Zfp366
|
zinc finger protein 366 |
| chr9_-_32452885 | 0.35 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr6_-_77956499 | 0.34 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr7_-_19556612 | 0.33 |
ENSMUST00000120537.8
|
Bcl3
|
B cell leukemia/lymphoma 3 |
| chr5_-_73496291 | 0.31 |
ENSMUST00000200830.4
ENSMUST00000201908.4 ENSMUST00000200776.4 ENSMUST00000087195.9 |
Ociad2
|
OCIA domain containing 2 |
| chr7_-_12002196 | 0.31 |
ENSMUST00000227973.2
ENSMUST00000228764.2 ENSMUST00000228482.2 ENSMUST00000227080.2 |
Vmn1r81
|
vomeronasal 1 receptor 81 |
| chr6_+_122803624 | 0.31 |
ENSMUST00000203075.2
|
Foxj2
|
forkhead box J2 |
| chr2_-_86940289 | 0.29 |
ENSMUST00000215828.3
|
Olfr259
|
olfactory receptor 259 |
| chr13_+_118851214 | 0.29 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr3_-_89820451 | 0.26 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr3_-_108819003 | 0.26 |
ENSMUST00000029480.9
|
Prpf38b
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B |
| chr17_+_37769807 | 0.26 |
ENSMUST00000214668.2
ENSMUST00000217602.2 ENSMUST00000214938.2 |
Olfr109
|
olfactory receptor 109 |
| chr5_-_90514061 | 0.26 |
ENSMUST00000081914.13
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr2_-_38177182 | 0.25 |
ENSMUST00000130472.8
|
Dennd1a
|
DENN/MADD domain containing 1A |
| chr5_-_66330394 | 0.24 |
ENSMUST00000201544.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr5_-_90514360 | 0.24 |
ENSMUST00000168058.7
ENSMUST00000014421.15 |
Ankrd17
|
ankyrin repeat domain 17 |
| chr5_+_117271632 | 0.21 |
ENSMUST00000179276.8
ENSMUST00000092889.12 ENSMUST00000145640.8 |
Taok3
|
TAO kinase 3 |
| chr5_-_116162415 | 0.21 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr9_+_38382403 | 0.21 |
ENSMUST00000214377.2
|
Olfr905
|
olfactory receptor 905 |
| chr9_+_118307250 | 0.21 |
ENSMUST00000111763.8
|
Eomes
|
eomesodermin |
| chr5_-_135378729 | 0.21 |
ENSMUST00000201784.4
ENSMUST00000201791.4 |
Fkbp6
|
FK506 binding protein 6 |
| chr5_-_135378896 | 0.21 |
ENSMUST00000201534.2
ENSMUST00000044972.11 |
Fkbp6
|
FK506 binding protein 6 |
| chr7_+_79675727 | 0.20 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr10_-_3217767 | 0.20 |
ENSMUST00000170893.3
|
H60c
|
histocompatibility 60c |
| chr5_-_90514034 | 0.19 |
ENSMUST00000218526.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr7_+_110371811 | 0.18 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr2_-_45003270 | 0.16 |
ENSMUST00000202935.4
ENSMUST00000068415.11 ENSMUST00000127520.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_36263531 | 0.16 |
ENSMUST00000072114.4
ENSMUST00000217511.2 |
Olfr338
|
olfactory receptor 338 |
| chr7_-_139662402 | 0.16 |
ENSMUST00000026541.15
ENSMUST00000211283.2 |
Caly
|
calcyon neuron-specific vesicular protein |
| chr11_-_97590460 | 0.16 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr19_-_6910195 | 0.16 |
ENSMUST00000236443.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr2_-_119378108 | 0.16 |
ENSMUST00000060009.9
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr5_-_65492940 | 0.16 |
ENSMUST00000203471.3
ENSMUST00000172732.8 ENSMUST00000204965.3 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr2_-_32976378 | 0.16 |
ENSMUST00000049618.9
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr16_+_91169671 | 0.15 |
ENSMUST00000023693.14
ENSMUST00000134491.9 ENSMUST00000117836.8 |
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr17_+_6157154 | 0.15 |
ENSMUST00000149756.8
|
Tulp4
|
tubby like protein 4 |
| chr10_-_127456791 | 0.15 |
ENSMUST00000118455.2
ENSMUST00000121829.8 |
Lrp1
|
low density lipoprotein receptor-related protein 1 |
| chr2_+_90912710 | 0.15 |
ENSMUST00000169852.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr14_-_46628033 | 0.14 |
ENSMUST00000074077.12
|
Bmp4
|
bone morphogenetic protein 4 |
| chr16_-_10884005 | 0.14 |
ENSMUST00000162323.2
|
Litaf
|
LPS-induced TN factor |
| chr18_-_75830595 | 0.14 |
ENSMUST00000165559.3
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr7_+_79676095 | 0.14 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr2_-_45002902 | 0.14 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_25066313 | 0.14 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chrX_+_41156752 | 0.13 |
ENSMUST00000126375.2
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr4_+_35152056 | 0.13 |
ENSMUST00000058595.7
|
Ifnk
|
interferon kappa |
| chr5_+_127709302 | 0.13 |
ENSMUST00000118139.3
|
Glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr18_+_73992446 | 0.12 |
ENSMUST00000120033.8
ENSMUST00000179472.8 ENSMUST00000119239.8 |
Mro
|
maestro |
| chr15_+_25940781 | 0.12 |
ENSMUST00000227275.2
|
Retreg1
|
reticulophagy regulator 1 |
| chrX_+_41156713 | 0.12 |
ENSMUST00000115094.8
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr10_+_19810037 | 0.12 |
ENSMUST00000095806.10
ENSMUST00000120259.8 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr16_+_91184661 | 0.11 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr14_-_20714634 | 0.11 |
ENSMUST00000119483.2
|
Synpo2l
|
synaptopodin 2-like |
| chr7_-_97970388 | 0.10 |
ENSMUST00000120520.8
|
Acer3
|
alkaline ceramidase 3 |
| chr3_+_122039206 | 0.10 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr16_+_15705141 | 0.10 |
ENSMUST00000096232.6
|
Cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr18_+_62086122 | 0.10 |
ENSMUST00000051720.6
ENSMUST00000235860.2 |
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr5_-_65492907 | 0.10 |
ENSMUST00000203581.3
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr1_+_170104889 | 0.10 |
ENSMUST00000179976.3
|
Sh2d1b1
|
SH2 domain containing 1B1 |
| chr10_+_84753480 | 0.10 |
ENSMUST00000038523.15
ENSMUST00000214693.2 ENSMUST00000095385.5 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr18_+_47054867 | 0.09 |
ENSMUST00000234910.2
|
Arl14epl
|
ADP-ribosylation factor-like 14 effector protein-like |
| chr16_+_91169770 | 0.09 |
ENSMUST00000089042.7
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr5_-_92511136 | 0.08 |
ENSMUST00000077820.6
|
Cxcl11
|
chemokine (C-X-C motif) ligand 11 |
| chrX_+_41157242 | 0.08 |
ENSMUST00000115095.9
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr7_+_110372860 | 0.08 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr1_+_44590791 | 0.08 |
ENSMUST00000160854.8
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr16_-_36486429 | 0.08 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr9_+_44394080 | 0.08 |
ENSMUST00000220303.2
|
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr5_+_123390149 | 0.08 |
ENSMUST00000121964.8
|
Wdr66
|
WD repeat domain 66 |
| chrX_-_166113457 | 0.08 |
ENSMUST00000145284.8
ENSMUST00000112164.2 ENSMUST00000137492.8 ENSMUST00000112161.8 ENSMUST00000060719.12 |
Tlr7
|
toll-like receptor 7 |
| chr4_+_145311722 | 0.08 |
ENSMUST00000105739.8
|
Zfp268
|
zinc finger protein 268 |
| chr12_-_118265163 | 0.08 |
ENSMUST00000221844.2
|
Sp4
|
trans-acting transcription factor 4 |
| chr17_-_35381945 | 0.08 |
ENSMUST00000174805.2
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr7_-_4535403 | 0.07 |
ENSMUST00000094897.5
|
Dnaaf3
|
dynein, axonemal assembly factor 3 |
| chr6_+_124470053 | 0.07 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr11_-_100650566 | 0.07 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chrX_+_72527208 | 0.07 |
ENSMUST00000033741.15
ENSMUST00000169489.2 |
Bgn
|
biglycan |
| chr6_+_38918327 | 0.07 |
ENSMUST00000160963.2
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr11_-_100650768 | 0.07 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr9_-_109168710 | 0.07 |
ENSMUST00000196351.2
ENSMUST00000200156.5 ENSMUST00000112040.9 ENSMUST00000112039.7 |
Fbxw28
|
F-box and WD-40 domain protein 28 |
| chr16_+_4412546 | 0.07 |
ENSMUST00000014447.13
|
Glis2
|
GLIS family zinc finger 2 |
| chr5_-_106606032 | 0.06 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr10_+_84591919 | 0.06 |
ENSMUST00000060397.13
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr17_-_24428351 | 0.06 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr17_-_24863907 | 0.06 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_-_140793990 | 0.06 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chr8_-_107620147 | 0.06 |
ENSMUST00000176090.2
|
Chtf8
|
CTF8, chromosome transmission fidelity factor 8 |
| chr2_+_181162278 | 0.06 |
ENSMUST00000072334.12
|
Dnajc5
|
DnaJ heat shock protein family (Hsp40) member C5 |
| chr1_-_135032972 | 0.05 |
ENSMUST00000044828.14
|
Lgr6
|
leucine-rich repeat-containing G protein-coupled receptor 6 |
| chr7_+_139673300 | 0.05 |
ENSMUST00000026540.9
|
Prap1
|
proline-rich acidic protein 1 |
| chr9_+_72439496 | 0.05 |
ENSMUST00000163401.9
ENSMUST00000093820.10 |
Rfx7
|
regulatory factor X, 7 |
| chr7_-_109215754 | 0.05 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chrX_-_8042129 | 0.05 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr7_-_4448180 | 0.04 |
ENSMUST00000138798.2
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr12_+_80992276 | 0.04 |
ENSMUST00000094693.11
|
Srsf5
|
serine and arginine-rich splicing factor 5 |
| chr2_+_79465761 | 0.04 |
ENSMUST00000111788.8
ENSMUST00000111784.9 |
Itprid2
|
ITPR interacting domain containing 2 |
| chr6_+_141194886 | 0.04 |
ENSMUST00000043259.10
|
Pde3a
|
phosphodiesterase 3A, cGMP inhibited |
| chr6_-_57668992 | 0.04 |
ENSMUST00000053386.6
|
Pyurf
|
Pigy upstream reading frame |
| chr14_-_55344004 | 0.04 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr2_+_92014451 | 0.04 |
ENSMUST00000111294.8
ENSMUST00000111293.9 |
Phf21a
|
PHD finger protein 21A |
| chr14_-_46627957 | 0.04 |
ENSMUST00000100676.3
|
Bmp4
|
bone morphogenetic protein 4 |
| chr13_+_42862957 | 0.04 |
ENSMUST00000066928.12
ENSMUST00000148891.8 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr19_+_37674029 | 0.04 |
ENSMUST00000073391.5
|
Cyp26c1
|
cytochrome P450, family 26, subfamily c, polypeptide 1 |
| chr7_+_34885782 | 0.04 |
ENSMUST00000135452.8
ENSMUST00000001854.12 |
Slc7a10
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 10 |
| chr4_+_145311759 | 0.04 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr2_+_155453103 | 0.04 |
ENSMUST00000092995.6
|
Myh7b
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr15_+_84052028 | 0.04 |
ENSMUST00000045289.6
|
Pnpla3
|
patatin-like phospholipase domain containing 3 |
| chr7_-_104426677 | 0.04 |
ENSMUST00000211384.2
ENSMUST00000053464.9 |
Usp17le
|
ubiquitin specific peptidase 17-like E |
| chr2_+_92014606 | 0.04 |
ENSMUST00000162146.8
ENSMUST00000111292.8 ENSMUST00000162497.8 |
Phf21a
|
PHD finger protein 21A |
| chr2_+_121787131 | 0.03 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr11_-_6470918 | 0.03 |
ENSMUST00000003459.4
|
Myo1g
|
myosin IG |
| chr4_+_140737955 | 0.03 |
ENSMUST00000071977.9
|
Mfap2
|
microfibrillar-associated protein 2 |
| chr4_+_40723083 | 0.03 |
ENSMUST00000149794.2
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr8_-_3770642 | 0.03 |
ENSMUST00000062037.7
|
Clec4g
|
C-type lectin domain family 4, member g |
| chr5_-_136227760 | 0.03 |
ENSMUST00000149151.2
ENSMUST00000151786.8 |
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr15_-_76010736 | 0.03 |
ENSMUST00000054022.12
ENSMUST00000089654.4 |
BC024139
|
cDNA sequence BC024139 |
| chr6_+_122490534 | 0.03 |
ENSMUST00000032210.14
ENSMUST00000148517.8 |
Mfap5
|
microfibrillar associated protein 5 |
| chr4_+_148686985 | 0.03 |
ENSMUST00000105701.9
ENSMUST00000052060.7 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr3_+_122039274 | 0.03 |
ENSMUST00000178826.8
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr2_-_88600832 | 0.02 |
ENSMUST00000217588.3
|
Olfr1200
|
olfactory receptor 1200 |
| chr17_-_24863956 | 0.02 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr4_-_150085722 | 0.02 |
ENSMUST00000153394.2
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr4_+_114914607 | 0.02 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr7_+_24561616 | 0.01 |
ENSMUST00000170837.3
|
Gm9844
|
predicted pseudogene 9844 |
| chr14_+_53497035 | 0.01 |
ENSMUST00000197614.2
|
Trav14n-2
|
T cell receptor alpha variable 14N-2 |
| chr15_-_54935535 | 0.01 |
ENSMUST00000041733.9
|
Taf2
|
TATA-box binding protein associated factor 2 |
| chrX_-_36432482 | 0.01 |
ENSMUST00000046557.6
|
Akap14
|
A kinase (PRKA) anchor protein 14 |
| chr5_-_73495895 | 0.01 |
ENSMUST00000202012.2
|
Ociad2
|
OCIA domain containing 2 |
| chr4_+_42655251 | 0.01 |
ENSMUST00000177785.3
|
Ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr4_+_109137561 | 0.01 |
ENSMUST00000177089.8
ENSMUST00000175776.8 ENSMUST00000132165.9 |
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr11_+_49419074 | 0.01 |
ENSMUST00000213707.2
ENSMUST00000213152.2 ENSMUST00000217564.2 ENSMUST00000213899.2 |
Olfr1382
|
olfactory receptor 1382 |
| chr9_+_44018583 | 0.01 |
ENSMUST00000152956.8
ENSMUST00000114815.3 ENSMUST00000206295.2 ENSMUST00000206769.2 ENSMUST00000205500.2 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr10_-_93375832 | 0.01 |
ENSMUST00000016034.3
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr14_+_53791444 | 0.01 |
ENSMUST00000198297.2
|
Trav14-1
|
T cell receptor alpha variable 14-1 |
| chr16_+_48104098 | 0.01 |
ENSMUST00000096045.9
ENSMUST00000050705.4 |
Dppa4
|
developmental pluripotency associated 4 |
| chr19_-_5452521 | 0.01 |
ENSMUST00000235569.2
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr7_-_25315299 | 0.01 |
ENSMUST00000098663.4
ENSMUST00000238895.2 |
Erich4
|
glutamate rich 4 |
| chr12_-_88290796 | 0.01 |
ENSMUST00000218054.3
|
Eif1ad15
|
eukaryotic translation initiation factor 1A domain containing 15 |
| chr13_-_99584091 | 0.01 |
ENSMUST00000223725.2
|
Map1b
|
microtubule-associated protein 1B |
| chr15_-_73056713 | 0.01 |
ENSMUST00000044113.12
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr2_+_79465696 | 0.01 |
ENSMUST00000111785.9
|
Itprid2
|
ITPR interacting domain containing 2 |
| chr9_-_109063792 | 0.01 |
ENSMUST00000197329.2
ENSMUST00000079548.12 |
Fbxw20
|
F-box and WD-40 domain protein 20 |
| chr11_-_5865124 | 0.01 |
ENSMUST00000109823.9
ENSMUST00000109822.8 |
Gck
|
glucokinase |
| chr9_+_44018551 | 0.01 |
ENSMUST00000114821.9
ENSMUST00000114818.9 |
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr6_+_4504814 | 0.01 |
ENSMUST00000141483.8
|
Col1a2
|
collagen, type I, alpha 2 |
| chr7_+_45199259 | 0.01 |
ENSMUST00000210797.2
|
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr11_-_53525520 | 0.01 |
ENSMUST00000020650.2
|
Il13
|
interleukin 13 |
| chr13_-_34027389 | 0.01 |
ENSMUST00000110275.8
ENSMUST00000145221.2 |
Serpinb6e
|
serine (or cysteine) peptidase inhibitor, clade B, member 6e |
| chr3_+_156267587 | 0.00 |
ENSMUST00000041425.12
ENSMUST00000106065.2 |
Negr1
|
neuronal growth regulator 1 |
| chr1_+_182591771 | 0.00 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr9_+_44018520 | 0.00 |
ENSMUST00000114816.8
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr9_-_109116739 | 0.00 |
ENSMUST00000112041.6
ENSMUST00000198844.5 |
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr4_-_41697040 | 0.00 |
ENSMUST00000102962.10
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr17_+_9068805 | 0.00 |
ENSMUST00000115720.8
|
Pde10a
|
phosphodiesterase 10A |
| chr4_+_42158092 | 0.00 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr13_-_4573312 | 0.00 |
ENSMUST00000221564.2
ENSMUST00000078239.5 ENSMUST00000080361.13 |
Akr1c20
|
aldo-keto reductase family 1, member C20 |
| chr9_-_109116150 | 0.00 |
ENSMUST00000198928.2
|
Fbxw14
|
F-box and WD-40 domain protein 14 |
| chr1_+_182591425 | 0.00 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr18_-_20192535 | 0.00 |
ENSMUST00000075214.9
ENSMUST00000039247.11 |
Dsc2
|
desmocollin 2 |
| chr10_+_24471340 | 0.00 |
ENSMUST00000020171.12
|
Ccn2
|
cellular communication network factor 2 |
| chr4_-_144134967 | 0.00 |
ENSMUST00000123854.2
ENSMUST00000030326.10 |
Pramel13
|
PRAME like 13 |
| chr2_-_121786573 | 0.00 |
ENSMUST00000104936.4
|
Mageb3
|
MAGE family member B3 |
| chr12_+_87561085 | 0.00 |
ENSMUST00000110152.3
|
Eif1ad8
|
eukaryotic translation initiation factor 1A domain containing 8 |
| chr2_+_158344553 | 0.00 |
ENSMUST00000109484.2
|
Adig
|
adipogenin |
| chr5_-_143211632 | 0.00 |
ENSMUST00000085733.9
|
Spdye4a
|
speedy/RINGO cell cycle regulator family, member E4A |
| chr2_+_158344532 | 0.00 |
ENSMUST00000059889.4
|
Adig
|
adipogenin |
| chr12_-_118265103 | 0.00 |
ENSMUST00000222314.2
ENSMUST00000026367.11 |
Sp4
|
trans-acting transcription factor 4 |
| chr19_+_29675224 | 0.00 |
ENSMUST00000025719.5
|
Mlana
|
melan-A |
| chr10_-_3217718 | 0.00 |
ENSMUST00000216211.2
|
H60c
|
histocompatibility 60c |
| chr7_-_108471586 | 0.00 |
ENSMUST00000216500.2
|
Olfr517
|
olfactory receptor 517 |
| chr4_+_3167320 | 0.00 |
ENSMUST00000226198.2
|
Vmn1r2
|
vomeronasal 1 receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1902623 | negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0061033 | bronchiole development(GO:0060435) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.3 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.1 | 0.3 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.2 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 | 0.5 | GO:1900020 | prolactin secretion(GO:0070459) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.4 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 1.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 2.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 3.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 2.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.6 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.4 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.1 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 3.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0070700 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |