Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
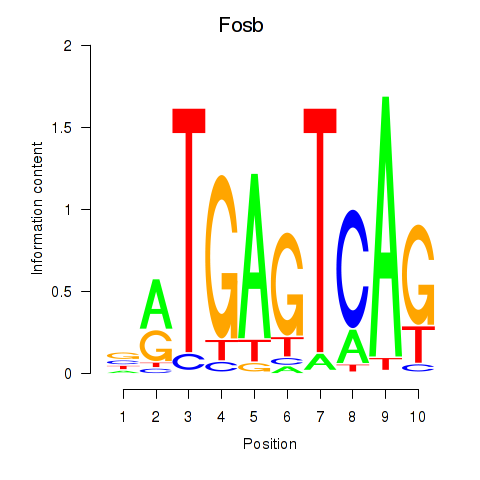
Results for Fosb
Z-value: 0.50
Transcription factors associated with Fosb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosb
|
ENSMUSG00000003545.4 | FBJ osteosarcoma oncogene B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fosb | mm39_v1_chr7_-_19043525_19043627 | -0.80 | 1.0e-01 | Click! |
Activity profile of Fosb motif
Sorted Z-values of Fosb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_42326714 | 0.48 |
ENSMUST00000203846.3
|
Zyx
|
zyxin |
| chr6_+_42326760 | 0.37 |
ENSMUST00000203652.3
ENSMUST00000070635.13 |
Zyx
|
zyxin |
| chr6_+_42326980 | 0.30 |
ENSMUST00000203849.2
|
Zyx
|
zyxin |
| chr10_-_85847697 | 0.30 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr6_+_42326934 | 0.29 |
ENSMUST00000203401.3
ENSMUST00000164375.4 |
Zyx
|
zyxin |
| chr16_-_4698148 | 0.29 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr18_+_82928782 | 0.28 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr2_-_18053595 | 0.25 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr7_-_80051455 | 0.24 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr17_+_36152383 | 0.23 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr7_-_29204812 | 0.23 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr3_-_131196213 | 0.23 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr12_-_84240781 | 0.22 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr8_-_70426910 | 0.22 |
ENSMUST00000116463.4
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr11_-_100288566 | 0.21 |
ENSMUST00000001592.15
ENSMUST00000107403.2 |
Jup
|
junction plakoglobin |
| chr5_-_62923463 | 0.19 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_-_124746510 | 0.18 |
ENSMUST00000149652.2
ENSMUST00000112476.8 ENSMUST00000004378.15 |
Eno2
|
enolase 2, gamma neuronal |
| chr9_-_32452885 | 0.18 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr19_-_32188413 | 0.18 |
ENSMUST00000151289.9
|
Sgms1
|
sphingomyelin synthase 1 |
| chr6_+_42326528 | 0.18 |
ENSMUST00000203329.3
|
Zyx
|
zyxin |
| chr8_+_4276827 | 0.17 |
ENSMUST00000053035.7
|
Lrrc8e
|
leucine rich repeat containing 8 family, member E |
| chr14_-_55995912 | 0.17 |
ENSMUST00000001497.9
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr11_-_69556904 | 0.16 |
ENSMUST00000018918.12
|
Cd68
|
CD68 antigen |
| chr6_+_86826470 | 0.16 |
ENSMUST00000089519.13
ENSMUST00000204414.3 |
Aak1
|
AP2 associated kinase 1 |
| chr1_-_182345011 | 0.15 |
ENSMUST00000068505.10
|
Capn2
|
calpain 2 |
| chr14_+_66872699 | 0.15 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr11_-_95966477 | 0.14 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_+_44151962 | 0.14 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr6_+_17463925 | 0.14 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr4_-_19922599 | 0.14 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr10_-_43880353 | 0.14 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr18_+_82928959 | 0.13 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr15_-_64184485 | 0.13 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr8_+_108020132 | 0.13 |
ENSMUST00000151114.8
ENSMUST00000125721.8 ENSMUST00000075922.11 |
Nfat5
|
nuclear factor of activated T cells 5 |
| chr11_-_69556888 | 0.13 |
ENSMUST00000108654.3
|
Cd68
|
CD68 antigen |
| chr6_+_49013601 | 0.12 |
ENSMUST00000204260.2
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr7_-_119461027 | 0.12 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr1_-_10790120 | 0.12 |
ENSMUST00000035577.7
|
Cpa6
|
carboxypeptidase A6 |
| chr12_+_71062733 | 0.12 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chrX_-_47551990 | 0.12 |
ENSMUST00000033429.9
ENSMUST00000140486.2 |
Elf4
|
E74-like factor 4 (ets domain transcription factor) |
| chr10_-_5872386 | 0.11 |
ENSMUST00000131996.8
ENSMUST00000064225.14 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr11_+_80319424 | 0.11 |
ENSMUST00000173938.8
ENSMUST00000017572.14 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr19_+_5927876 | 0.10 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr11_-_102255999 | 0.10 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr17_+_78815493 | 0.10 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr12_+_83678987 | 0.09 |
ENSMUST00000048155.16
ENSMUST00000182618.8 ENSMUST00000183154.8 ENSMUST00000182036.8 ENSMUST00000182347.8 |
Rbm25
|
RNA binding motif protein 25 |
| chr3_-_79439181 | 0.09 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr5_+_32293145 | 0.08 |
ENSMUST00000031017.11
|
Fosl2
|
fos-like antigen 2 |
| chr17_+_48080113 | 0.08 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr10_-_5872341 | 0.08 |
ENSMUST00000117676.8
ENSMUST00000019909.8 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr9_-_108888779 | 0.08 |
ENSMUST00000061973.5
|
Trex1
|
three prime repair exonuclease 1 |
| chr7_-_4781140 | 0.07 |
ENSMUST00000094892.12
|
Il11
|
interleukin 11 |
| chr5_-_93192881 | 0.07 |
ENSMUST00000061328.6
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr12_+_111132908 | 0.07 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr15_-_75886166 | 0.07 |
ENSMUST00000060807.12
|
Fam83h
|
family with sequence similarity 83, member H |
| chr2_+_118493713 | 0.07 |
ENSMUST00000099557.10
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr12_+_16944896 | 0.07 |
ENSMUST00000020904.8
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2 |
| chr7_+_126376099 | 0.06 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr10_-_121422635 | 0.05 |
ENSMUST00000219493.2
ENSMUST00000020316.4 |
Tbk1
|
TANK-binding kinase 1 |
| chr8_+_108020092 | 0.05 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr3_-_95139472 | 0.05 |
ENSMUST00000196025.5
ENSMUST00000198948.5 |
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr5_-_5315968 | 0.05 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr19_-_4189603 | 0.04 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
| chr4_-_140501507 | 0.04 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr15_-_74599860 | 0.04 |
ENSMUST00000023261.4
ENSMUST00000190433.2 |
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr2_+_51928017 | 0.03 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr17_-_24863907 | 0.03 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_+_139414057 | 0.03 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr2_-_90900628 | 0.03 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr11_-_120358239 | 0.03 |
ENSMUST00000076921.7
|
Arl16
|
ADP-ribosylation factor-like 16 |
| chr10_-_63926044 | 0.03 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr9_-_108888284 | 0.03 |
ENSMUST00000112053.2
|
Trex1
|
three prime repair exonuclease 1 |
| chr11_+_51541728 | 0.03 |
ENSMUST00000117859.8
ENSMUST00000064493.6 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr19_-_38113249 | 0.02 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr11_-_100139728 | 0.02 |
ENSMUST00000007280.9
|
Krt16
|
keratin 16 |
| chr6_+_53264255 | 0.02 |
ENSMUST00000203528.3
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr18_+_4994600 | 0.02 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr17_-_24863956 | 0.02 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr10_-_18619439 | 0.02 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chr19_+_5497575 | 0.02 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr11_+_61575245 | 0.02 |
ENSMUST00000093019.6
|
Fam83g
|
family with sequence similarity 83, member G |
| chr16_+_17379749 | 0.02 |
ENSMUST00000171002.10
ENSMUST00000023441.11 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr1_-_75240551 | 0.02 |
ENSMUST00000186178.7
ENSMUST00000189769.7 ENSMUST00000027404.12 |
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr13_+_30843937 | 0.01 |
ENSMUST00000091672.13
|
Dusp22
|
dual specificity phosphatase 22 |
| chr7_-_141926079 | 0.01 |
ENSMUST00000059223.15
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr9_+_122717536 | 0.01 |
ENSMUST00000063980.8
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr7_+_75879603 | 0.01 |
ENSMUST00000156166.8
|
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr1_-_74932266 | 0.01 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr19_+_4189786 | 0.01 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr15_+_102412157 | 0.01 |
ENSMUST00000096145.5
|
Gm10337
|
predicted gene 10337 |
| chr11_-_69439934 | 0.01 |
ENSMUST00000108659.2
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr11_+_4207557 | 0.00 |
ENSMUST00000066283.12
|
Lif
|
leukemia inhibitory factor |
| chr7_+_6048160 | 0.00 |
ENSMUST00000037728.13
ENSMUST00000121583.2 |
Nlrp4c
|
NLR family, pyrin domain containing 4C |
| chr3_-_92481033 | 0.00 |
ENSMUST00000053107.6
|
Ivl
|
involucrin |
| chr5_+_31274064 | 0.00 |
ENSMUST00000202769.2
|
Trim54
|
tripartite motif-containing 54 |
| chr5_+_31274046 | 0.00 |
ENSMUST00000013771.15
|
Trim54
|
tripartite motif-containing 54 |
| chr19_-_38113056 | 0.00 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr18_-_31450095 | 0.00 |
ENSMUST00000139924.2
ENSMUST00000153060.8 |
Rit2
|
Ras-like without CAAX 2 |
| chr6_-_124746445 | 0.00 |
ENSMUST00000156033.2
|
Eno2
|
enolase 2, gamma neuronal |
| chr7_+_140659038 | 0.00 |
ENSMUST00000159375.8
|
Pkp3
|
plakophilin 3 |
| chr4_+_42158092 | 0.00 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr4_+_109263820 | 0.00 |
ENSMUST00000124209.8
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr3_+_102377234 | 0.00 |
ENSMUST00000035952.5
ENSMUST00000198168.5 ENSMUST00000106925.9 |
Ngf
|
nerve growth factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |