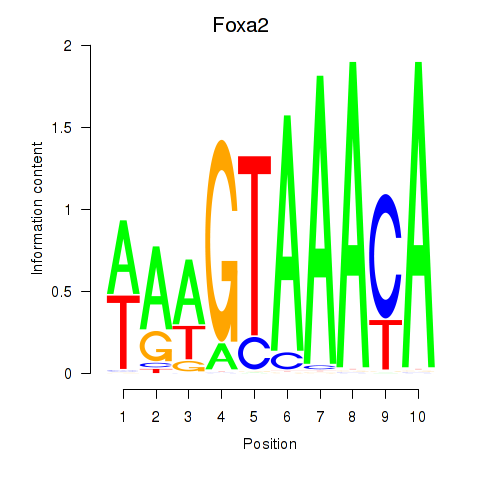
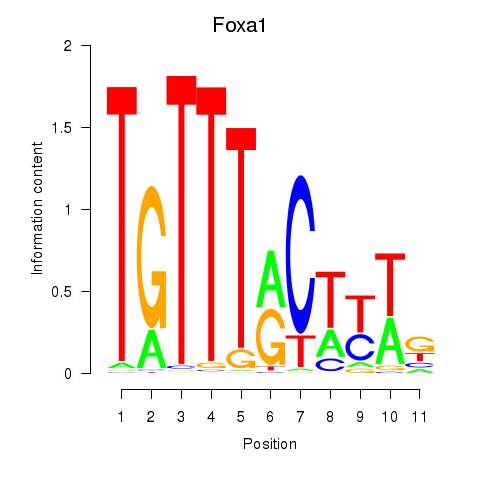
|
chr7_+_43000765
Show fit
|
0.73 |
ENSMUST00000012798.14
ENSMUST00000122423.8
|
Siglecf
|
sialic acid binding Ig-like lectin F
|
|
chr7_+_43000838
Show fit
|
0.64 |
ENSMUST00000206299.2
ENSMUST00000121494.2
|
Siglecf
|
sialic acid binding Ig-like lectin F
|
|
chr13_+_23715220
Show fit
|
0.33 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8
|
|
chr19_+_23118545
Show fit
|
0.32 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9
|
|
chr2_+_158148413
Show fit
|
0.28 |
ENSMUST00000109491.8
ENSMUST00000016168.9
|
Lbp
|
lipopolysaccharide binding protein
|
|
chr5_+_16758777
Show fit
|
0.28 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor
|
|
chr5_+_16758538
Show fit
|
0.27 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor
|
|
chr5_+_16758897
Show fit
|
0.25 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor
|
|
chr16_-_95387444
Show fit
|
0.24 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor
|
|
chr16_-_4698148
Show fit
|
0.22 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1
|
|
chr12_+_77284822
Show fit
|
0.21 |
ENSMUST00000177595.9
ENSMUST00000171770.10
|
Fut8
|
fucosyltransferase 8
|
|
chr9_-_103569984
Show fit
|
0.21 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108
|
|
chr11_+_108814007
Show fit
|
0.20 |
ENSMUST00000106711.2
|
Axin2
|
axin 2
|
|
chr5_+_102629240
Show fit
|
0.20 |
ENSMUST00000073302.12
ENSMUST00000094559.9
|
Arhgap24
|
Rho GTPase activating protein 24
|
|
chr5_+_102629365
Show fit
|
0.19 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24
|
|
chr2_-_18053158
Show fit
|
0.19 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1
|
|
chr12_-_31549538
Show fit
|
0.19 |
ENSMUST00000064240.14
ENSMUST00000185739.8
ENSMUST00000188326.3
ENSMUST00000101499.10
ENSMUST00000085487.12
|
Cbll1
|
Casitas B-lineage lymphoma-like 1
|
|
chr10_+_128744689
Show fit
|
0.18 |
ENSMUST00000105229.9
|
Cd63
|
CD63 antigen
|
|
chr6_-_86742847
Show fit
|
0.18 |
ENSMUST00000113675.8
|
Anxa4
|
annexin A4
|
|
chrM_+_2743
Show fit
|
0.18 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1
|
|
chr6_-_86742789
Show fit
|
0.18 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4
|
|
chr14_+_73475335
Show fit
|
0.17 |
ENSMUST00000044405.8
|
Lpar6
|
lysophosphatidic acid receptor 6
|
|
chr14_+_20732804
Show fit
|
0.17 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae)
|
|
chr1_-_69726384
Show fit
|
0.17 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2
|
|
chr12_+_112645237
Show fit
|
0.17 |
ENSMUST00000174780.2
ENSMUST00000169593.2
ENSMUST00000173942.2
|
Zbtb42
|
zinc finger and BTB domain containing 42
|
|
chr7_-_3551003
Show fit
|
0.16 |
ENSMUST00000065703.9
ENSMUST00000203020.3
ENSMUST00000203821.3
|
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1
|
|
chr3_-_131138541
Show fit
|
0.16 |
ENSMUST00000090246.5
ENSMUST00000126569.2
|
Sgms2
|
sphingomyelin synthase 2
|
|
chr2_-_170269748
Show fit
|
0.15 |
ENSMUST00000013667.3
ENSMUST00000109152.9
ENSMUST00000068137.11
|
Bcas1
|
breast carcinoma amplified sequence 1
|
|
chr2_-_45001141
Show fit
|
0.15 |
ENSMUST00000201969.4
ENSMUST00000201623.4
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr12_-_32000169
Show fit
|
0.15 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1
|
|
chr3_+_96127174
Show fit
|
0.14 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21
|
|
chr2_-_18053595
Show fit
|
0.14 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1
|
|
chr8_+_66838927
Show fit
|
0.14 |
ENSMUST00000039540.12
ENSMUST00000110253.3
|
Marchf1
|
membrane associated ring-CH-type finger 1
|
|
chr15_-_66432938
Show fit
|
0.14 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71
|
|
chr1_+_157353696
Show fit
|
0.13 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae)
|
|
chr10_+_111342147
Show fit
|
0.11 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1
|
|
chr3_-_143908111
Show fit
|
0.11 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4
|
|
chr7_+_119499322
Show fit
|
0.11 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1
|
|
chr8_+_70826191
Show fit
|
0.11 |
ENSMUST00000003659.9
|
Comp
|
cartilage oligomeric matrix protein
|
|
chr10_+_23727325
Show fit
|
0.11 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3
|
|
chr5_-_124003553
Show fit
|
0.10 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2
|
|
chr17_+_69144053
Show fit
|
0.10 |
ENSMUST00000178545.3
|
Tmem200c
|
transmembrane protein 200C
|
|
chr5_-_3707166
Show fit
|
0.10 |
ENSMUST00000196304.2
|
Gatad1
|
GATA zinc finger domain containing 1
|
|
chr2_-_51862941
Show fit
|
0.09 |
ENSMUST00000145481.8
ENSMUST00000112705.9
|
Nmi
|
N-myc (and STAT) interactor
|
|
chr13_+_38224201
Show fit
|
0.09 |
ENSMUST00000224956.2
|
Riok1
|
RIO kinase 1
|
|
chr3_-_143908060
Show fit
|
0.09 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4
|
|
chr9_+_86625694
Show fit
|
0.08 |
ENSMUST00000179574.2
ENSMUST00000036426.13
|
Prss35
|
protease, serine 35
|
|
chr19_-_37184692
Show fit
|
0.08 |
ENSMUST00000132580.8
ENSMUST00000079754.11
ENSMUST00000136286.8
ENSMUST00000126188.8
ENSMUST00000126781.2
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chrX_+_158491589
Show fit
|
0.08 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr17_-_34847354
Show fit
|
0.08 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2
|
|
chr12_-_32000209
Show fit
|
0.08 |
ENSMUST00000176084.2
ENSMUST00000176103.8
ENSMUST00000167458.9
|
Hbp1
|
high mobility group box transcription factor 1
|
|
chr10_+_89580849
Show fit
|
0.07 |
ENSMUST00000020112.7
|
Uhrf1bp1l
|
UHRF1 (ICBP90) binding protein 1-like
|
|
chr9_-_53447908
Show fit
|
0.07 |
ENSMUST00000150244.2
|
Atm
|
ataxia telangiectasia mutated
|
|
chr6_+_90596123
Show fit
|
0.07 |
ENSMUST00000032177.10
|
Slc41a3
|
solute carrier family 41, member 3
|
|
chr5_+_31771281
Show fit
|
0.07 |
ENSMUST00000031024.14
ENSMUST00000202033.2
|
Mrpl33
Gm43809
|
mitochondrial ribosomal protein L33
predicted gene 43809
|
|
chr15_-_50753792
Show fit
|
0.07 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1
|
|
chr10_-_21036792
Show fit
|
0.07 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene
|
|
chr3_+_137573839
Show fit
|
0.07 |
ENSMUST00000197711.2
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14
|
|
chr11_+_70323452
Show fit
|
0.07 |
ENSMUST00000084954.13
ENSMUST00000108568.10
ENSMUST00000079056.9
ENSMUST00000102564.11
ENSMUST00000124943.8
ENSMUST00000150076.8
ENSMUST00000102563.2
|
Arrb2
|
arrestin, beta 2
|
|
chr10_-_21036824
Show fit
|
0.07 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene
|
|
chr17_+_25097199
Show fit
|
0.07 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit
|
|
chr11_+_33997114
Show fit
|
0.06 |
ENSMUST00000109329.9
|
Lcp2
|
lymphocyte cytosolic protein 2
|
|
chr10_+_128745214
Show fit
|
0.06 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen
|
|
chr1_-_128520002
Show fit
|
0.06 |
ENSMUST00000052172.7
ENSMUST00000142893.2
|
Cxcr4
|
chemokine (C-X-C motif) receptor 4
|
|
chr17_+_47815998
Show fit
|
0.06 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3
|
|
chr6_-_52160816
Show fit
|
0.06 |
ENSMUST00000134831.2
|
Hoxa3
|
homeobox A3
|
|
chr2_-_70655997
Show fit
|
0.06 |
ENSMUST00000038584.9
|
Tlk1
|
tousled-like kinase 1
|
|
chr4_+_133788065
Show fit
|
0.06 |
ENSMUST00000227683.2
|
Crybg2
|
crystallin beta-gamma domain containing 2
|
|
chr11_-_70350783
Show fit
|
0.06 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16
|
|
chr9_-_71070506
Show fit
|
0.06 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9
|
|
chr5_-_66238313
Show fit
|
0.06 |
ENSMUST00000202700.4
ENSMUST00000094757.9
ENSMUST00000113724.6
|
Rbm47
|
RNA binding motif protein 47
|
|
chr17_+_47816042
Show fit
|
0.06 |
ENSMUST00000183044.8
ENSMUST00000037333.17
|
Ccnd3
|
cyclin D3
|
|
chr18_-_60635059
Show fit
|
0.06 |
ENSMUST00000042710.8
|
Smim3
|
small integral membrane protein 3
|
|
chr10_+_57521930
Show fit
|
0.06 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific
|
|
chr5_-_66309244
Show fit
|
0.06 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47
|
|
chr2_+_69553141
Show fit
|
0.06 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G)
|
|
chr9_+_107457316
Show fit
|
0.05 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit
|
|
chr2_+_110427643
Show fit
|
0.05 |
ENSMUST00000045972.13
ENSMUST00000111026.3
|
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr10_-_128016135
Show fit
|
0.05 |
ENSMUST00000238843.2
ENSMUST00000099139.9
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr2_-_51863203
Show fit
|
0.05 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor
|
|
chr2_-_73722874
Show fit
|
0.05 |
ENSMUST00000136958.8
ENSMUST00000112010.9
ENSMUST00000128531.8
ENSMUST00000112017.8
|
Atf2
|
activating transcription factor 2
|
|
chr10_+_57521958
Show fit
|
0.05 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific
|
|
chr11_-_75918551
Show fit
|
0.05 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B
|
|
chr2_-_45000389
Show fit
|
0.05 |
ENSMUST00000201804.4
ENSMUST00000028229.13
ENSMUST00000202187.4
ENSMUST00000153561.6
ENSMUST00000201490.2
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr8_-_32408864
Show fit
|
0.05 |
ENSMUST00000073884.7
ENSMUST00000238812.2
|
Nrg1
|
neuregulin 1
|
|
chr7_-_80053063
Show fit
|
0.05 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr2_+_69553384
Show fit
|
0.05 |
ENSMUST00000040915.15
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G)
|
|
chr7_+_141392802
Show fit
|
0.05 |
ENSMUST00000165147.3
|
Muc5b
|
mucin 5, subtype B, tracheobronchial
|
|
chr13_-_74956030
Show fit
|
0.05 |
ENSMUST00000065629.6
|
Cast
|
calpastatin
|
|
chr2_-_111779785
Show fit
|
0.05 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307
|
|
chr9_+_78522783
Show fit
|
0.05 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen
|
|
chr12_+_55211069
Show fit
|
0.05 |
ENSMUST00000218889.2
|
Srp54b
|
signal recognition particle 54B
|
|
chr3_+_88486921
Show fit
|
0.04 |
ENSMUST00000193934.6
ENSMUST00000192495.6
ENSMUST00000035785.9
|
Ssr2
|
signal sequence receptor, beta
|
|
chr2_-_168443540
Show fit
|
0.04 |
ENSMUST00000171689.8
ENSMUST00000137451.2
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2
|
|
chr5_+_25452342
Show fit
|
0.04 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11
|
|
chr2_-_90309509
Show fit
|
0.04 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J
|
|
chrX_+_158086253
Show fit
|
0.04 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3
|
|
chr1_-_182929025
Show fit
|
0.04 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1
|
|
chr2_+_30485048
Show fit
|
0.04 |
ENSMUST00000102853.4
|
Cstad
|
CSA-conditional, T cell activation-dependent protein
|
|
chr11_+_85243970
Show fit
|
0.04 |
ENSMUST00000108056.8
ENSMUST00000108061.8
ENSMUST00000108062.8
ENSMUST00000138423.8
ENSMUST00000092821.10
ENSMUST00000074875.11
|
Bcas3
|
breast carcinoma amplified sequence 3
|
|
chr15_+_59520493
Show fit
|
0.04 |
ENSMUST00000118228.2
|
Trib1
|
tribbles pseudokinase 1
|
|
chr15_+_3300249
Show fit
|
0.04 |
ENSMUST00000082424.12
ENSMUST00000159158.9
ENSMUST00000159216.10
ENSMUST00000160311.3
|
Selenop
|
selenoprotein P
|
|
chr6_+_14901343
Show fit
|
0.04 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2
|
|
chr17_-_45125468
Show fit
|
0.04 |
ENSMUST00000159943.8
ENSMUST00000160673.8
|
Runx2
|
runt related transcription factor 2
|
|
chr10_-_67120959
Show fit
|
0.04 |
ENSMUST00000159002.2
ENSMUST00000077839.13
|
Nrbf2
|
nuclear receptor binding factor 2
|
|
chr4_+_118266526
Show fit
|
0.04 |
ENSMUST00000084319.11
ENSMUST00000106384.10
ENSMUST00000126089.8
ENSMUST00000073881.8
ENSMUST00000019229.15
|
Med8
|
mediator complex subunit 8
|
|
chr3_-_27765381
Show fit
|
0.04 |
ENSMUST00000193779.2
|
Fndc3b
|
fibronectin type III domain containing 3B
|
|
chr6_-_13677928
Show fit
|
0.04 |
ENSMUST00000203078.2
ENSMUST00000045235.8
|
Bmt2
|
base methyltransferase of 25S rRNA 2
|
|
chr2_-_163486998
Show fit
|
0.04 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3
|
|
chr16_+_37688744
Show fit
|
0.04 |
ENSMUST00000078717.7
|
Lrrc58
|
leucine rich repeat containing 58
|
|
chrX_+_93278526
Show fit
|
0.04 |
ENSMUST00000113908.8
ENSMUST00000113916.10
|
Klhl15
|
kelch-like 15
|
|
chr15_-_80989200
Show fit
|
0.04 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A
|
|
chr17_+_8144822
Show fit
|
0.04 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein
|
|
chr7_+_78563513
Show fit
|
0.04 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein
|
|
chr9_+_53448322
Show fit
|
0.03 |
ENSMUST00000035850.8
|
Npat
|
nuclear protein in the AT region
|
|
chr7_+_45370607
Show fit
|
0.03 |
ENSMUST00000129507.5
|
Fam83e
|
family with sequence similarity 83, member E
|
|
chr2_+_109848224
Show fit
|
0.03 |
ENSMUST00000150183.9
|
Ccdc34
|
coiled-coil domain containing 34
|
|
chr18_-_7626808
Show fit
|
0.03 |
ENSMUST00000234812.2
ENSMUST00000235093.2
ENSMUST00000234571.2
ENSMUST00000115869.4
|
Mpp7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7)
|
|
chr15_-_102630589
Show fit
|
0.03 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1
|
|
chr14_+_33193765
Show fit
|
0.03 |
ENSMUST00000208577.2
|
Frmpd2
|
FERM and PDZ domain containing 2
|
|
chr1_+_43484895
Show fit
|
0.03 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2
|
|
chr4_+_150938376
Show fit
|
0.03 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1
|
|
chr19_+_26600820
Show fit
|
0.03 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr11_+_31950452
Show fit
|
0.03 |
ENSMUST00000109409.8
ENSMUST00000020537.9
|
Nsg2
|
neuron specific gene family member 2
|
|
chr19_-_9065309
Show fit
|
0.03 |
ENSMUST00000025554.3
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr1_-_97904958
Show fit
|
0.03 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr17_+_71511642
Show fit
|
0.03 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2
|
|
chr17_+_73144531
Show fit
|
0.03 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5
|
|
chr4_-_102883905
Show fit
|
0.03 |
ENSMUST00000084382.6
ENSMUST00000106869.3
|
Insl5
|
insulin-like 5
|
|
chr19_-_6964988
Show fit
|
0.03 |
ENSMUST00000130048.8
ENSMUST00000025914.7
|
Vegfb
|
vascular endothelial growth factor B
|
|
chr14_-_55880708
Show fit
|
0.03 |
ENSMUST00000120041.8
ENSMUST00000121937.8
ENSMUST00000133707.2
ENSMUST00000002391.15
ENSMUST00000121791.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1
|
|
chr10_-_78554104
Show fit
|
0.03 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14
|
|
chr8_+_88978600
Show fit
|
0.03 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B
|
|
chr5_+_115080217
Show fit
|
0.03 |
ENSMUST00000031538.9
|
2210016L21Rik
|
RIKEN cDNA 2210016L21 gene
|
|
chr19_-_12773472
Show fit
|
0.03 |
ENSMUST00000038627.9
|
Zfp91
|
zinc finger protein 91
|
|
chr4_+_42922253
Show fit
|
0.03 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24
|
|
chr6_-_53955647
Show fit
|
0.03 |
ENSMUST00000204674.3
ENSMUST00000166545.2
ENSMUST00000203101.3
|
Cpvl
|
carboxypeptidase, vitellogenic-like
|
|
chr11_+_29413734
Show fit
|
0.03 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A
|
|
chr10_+_96453408
Show fit
|
0.03 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1
|
|
chr9_-_69668204
Show fit
|
0.03 |
ENSMUST00000071281.5
|
Foxb1
|
forkhead box B1
|
|
chr18_-_88945571
Show fit
|
0.03 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6
|
|
chr2_-_90735171
Show fit
|
0.03 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3
|
|
chr2_+_18069375
Show fit
|
0.02 |
ENSMUST00000114671.8
ENSMUST00000114680.9
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10
|
|
chrX_-_107877909
Show fit
|
0.02 |
ENSMUST00000101283.4
ENSMUST00000150434.8
|
Brwd3
|
bromodomain and WD repeat domain containing 3
|
|
chr2_-_160701523
Show fit
|
0.02 |
ENSMUST00000103112.8
|
Zhx3
|
zinc fingers and homeoboxes 3
|
|
chr1_+_130793406
Show fit
|
0.02 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor
|
|
chr9_-_123778334
Show fit
|
0.02 |
ENSMUST00000071404.5
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1
|
|
chr12_+_56742413
Show fit
|
0.02 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9
|
|
chr15_-_84739292
Show fit
|
0.02 |
ENSMUST00000016768.12
|
Phf21b
|
PHD finger protein 21B
|
|
chr8_+_107329983
Show fit
|
0.02 |
ENSMUST00000000312.12
ENSMUST00000167688.2
|
Cdh1
|
cadherin 1
|
|
chr11_+_33996920
Show fit
|
0.02 |
ENSMUST00000052413.12
|
Lcp2
|
lymphocyte cytosolic protein 2
|
|
chr17_+_47815968
Show fit
|
0.02 |
ENSMUST00000182129.8
ENSMUST00000171031.8
|
Ccnd3
|
cyclin D3
|
|
chr15_+_54274151
Show fit
|
0.02 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10
|
|
chr3_-_102871440
Show fit
|
0.02 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5
|
|
chr16_-_22475915
Show fit
|
0.02 |
ENSMUST00000089925.10
|
Dgkg
|
diacylglycerol kinase, gamma
|
|
chr8_+_94537910
Show fit
|
0.02 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O
|
|
chr3_+_69129745
Show fit
|
0.02 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14
|
|
chr5_+_43672856
Show fit
|
0.02 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7
|
|
chr1_-_173018204
Show fit
|
0.02 |
ENSMUST00000215878.2
ENSMUST00000201132.3
|
Olfr1406
|
olfactory receptor 1406
|
|
chr3_-_133250889
Show fit
|
0.02 |
ENSMUST00000197118.5
|
Tet2
|
tet methylcytosine dioxygenase 2
|
|
chr6_+_129568745
Show fit
|
0.02 |
ENSMUST00000032268.14
ENSMUST00000112063.9
ENSMUST00000119520.8
|
Klrd1
|
killer cell lectin-like receptor, subfamily D, member 1
|
|
chr6_-_40562700
Show fit
|
0.02 |
ENSMUST00000177178.2
ENSMUST00000129948.9
ENSMUST00000101491.11
|
Clec5a
|
C-type lectin domain family 5, member a
|
|
chr3_-_52012462
Show fit
|
0.02 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3
|
|
chr11_+_78389913
Show fit
|
0.02 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin
|
|
chr14_-_110992533
Show fit
|
0.02 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6
|
|
chr7_-_44498305
Show fit
|
0.02 |
ENSMUST00000207293.2
ENSMUST00000207532.2
|
Tbc1d17
|
TBC1 domain family, member 17
|
|
chr11_+_96820220
Show fit
|
0.02 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like
|
|
chr9_+_53757448
Show fit
|
0.02 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin
|
|
chr10_-_61814852
Show fit
|
0.02 |
ENSMUST00000105453.8
ENSMUST00000105452.9
ENSMUST00000105454.3
|
Col13a1
|
collagen, type XIII, alpha 1
|
|
chr1_-_87501548
Show fit
|
0.02 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor
|
|
chr15_+_5215180
Show fit
|
0.02 |
ENSMUST00000081640.12
|
Ttc33
|
tetratricopeptide repeat domain 33
|
|
chrX_+_93278203
Show fit
|
0.02 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15
|
|
chr9_+_32135540
Show fit
|
0.02 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr14_-_88708782
Show fit
|
0.02 |
ENSMUST00000192557.2
ENSMUST00000061628.7
|
Pcdh20
|
protocadherin 20
|
|
chr18_-_60781365
Show fit
|
0.02 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin
|
|
chr16_-_74208180
Show fit
|
0.02 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2
|
|
chr17_+_12597490
Show fit
|
0.02 |
ENSMUST00000014578.7
|
Plg
|
plasminogen
|
|
chr5_+_86219593
Show fit
|
0.02 |
ENSMUST00000198435.5
ENSMUST00000031171.9
|
Stap1
|
signal transducing adaptor family member 1
|
|
chr7_+_104315960
Show fit
|
0.02 |
ENSMUST00000214986.2
|
Olfr659
|
olfactory receptor 659
|
|
chr7_+_106692427
Show fit
|
0.02 |
ENSMUST00000210568.4
|
Olfr17
|
olfactory receptor 17
|
|
chr16_-_96971905
Show fit
|
0.02 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule
|
|
chr18_+_37085673
Show fit
|
0.02 |
ENSMUST00000192512.6
ENSMUST00000192295.2
ENSMUST00000115661.5
|
Pcdha4
Gm42416
|
protocadherin alpha 4
predicted gene, 42416
|
|
chr6_+_128991064
Show fit
|
0.02 |
ENSMUST00000204981.2
|
Clec2f
|
C-type lectin domain family 2, member f
|
|
chr4_-_57916283
Show fit
|
0.02 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene
|
|
chr8_-_62576140
Show fit
|
0.02 |
ENSMUST00000034052.14
ENSMUST00000034054.9
|
Anxa10
|
annexin A10
|
|
chr17_-_24382638
Show fit
|
0.02 |
ENSMUST00000129523.3
ENSMUST00000138685.3
ENSMUST00000040735.12
|
Amdhd2
|
amidohydrolase domain containing 2
|
|
chr6_+_97968737
Show fit
|
0.02 |
ENSMUST00000043628.13
ENSMUST00000203938.2
|
Mitf
|
melanogenesis associated transcription factor
|
|
chrX_-_42256694
Show fit
|
0.02 |
ENSMUST00000115058.8
ENSMUST00000115059.8
|
Tenm1
|
teneurin transmembrane protein 1
|
|
chr18_+_22478146
Show fit
|
0.02 |
ENSMUST00000120223.8
ENSMUST00000097655.4
|
Asxl3
|
ASXL transcriptional regulator 3
|
|
chr8_-_23143422
Show fit
|
0.02 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta
|
|
chr15_+_10177709
Show fit
|
0.02 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor
|
|
chr2_+_80447389
Show fit
|
0.02 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19
|
|
chr10_+_129334519
Show fit
|
0.02 |
ENSMUST00000215067.2
|
Olfr790
|
olfactory receptor 790
|
|
chr9_+_35819708
Show fit
|
0.02 |
ENSMUST00000176049.2
ENSMUST00000176153.2
|
Pate13
|
prostate and testis expressed 13
|
|
chr7_-_85895409
Show fit
|
0.02 |
ENSMUST00000165771.2
ENSMUST00000233075.2
ENSMUST00000233317.2
ENSMUST00000233312.2
ENSMUST00000233928.2
ENSMUST00000232799.2
ENSMUST00000233744.2
|
Vmn2r76
|
vomeronasal 2, receptor 76
|
|
chr3_+_130906434
Show fit
|
0.02 |
ENSMUST00000098611.4
|
Lef1
|
lymphoid enhancer binding factor 1
|
|
chr11_+_115225557
Show fit
|
0.02 |
ENSMUST00000106543.8
ENSMUST00000019006.5
|
Otop3
|
otopetrin 3
|
|
chr2_-_62313981
Show fit
|
0.02 |
ENSMUST00000136686.2
ENSMUST00000102733.10
|
Gcg
|
glucagon
|
|
chr7_+_19093665
Show fit
|
0.02 |
ENSMUST00000140836.8
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr8_-_32408380
Show fit
|
0.02 |
ENSMUST00000208497.3
ENSMUST00000207584.3
|
Nrg1
|
neuregulin 1
|