Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Foxa3
Z-value: 1.11
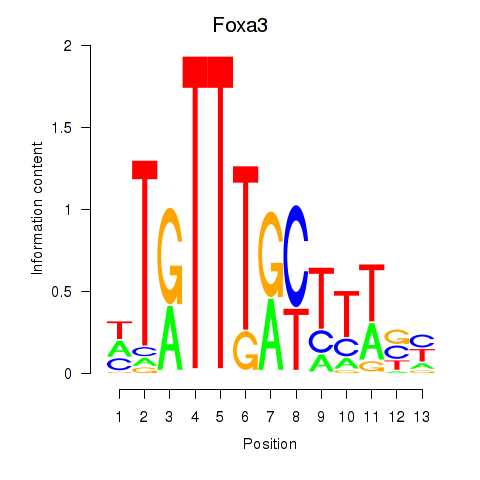
Transcription factors associated with Foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxa3
|
ENSMUSG00000040891.7 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxa3 | mm39_v1_chr7_-_18757461_18757477 | 0.17 | 7.8e-01 | Click! |
Activity profile of Foxa3 motif
Sorted Z-values of Foxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23715220 | 2.32 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_+_23922783 | 1.31 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chr16_+_35977170 | 1.20 |
ENSMUST00000079184.6
|
Stfa2l1
|
stefin A2 like 1 |
| chr7_-_3551003 | 1.11 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr2_+_84810802 | 0.91 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr14_+_51366306 | 0.76 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr14_+_51366512 | 0.72 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr3_+_96127174 | 0.70 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr2_-_90309509 | 0.64 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr1_+_32211792 | 0.59 |
ENSMUST00000027226.12
ENSMUST00000189878.2 ENSMUST00000188257.7 ENSMUST00000185666.2 |
Khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr2_-_111779785 | 0.51 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr9_+_106247943 | 0.50 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr4_-_55532453 | 0.47 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr9_+_107457316 | 0.45 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr1_+_165591315 | 0.43 |
ENSMUST00000111432.10
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr15_+_3300249 | 0.42 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr6_-_99005212 | 0.40 |
ENSMUST00000177437.8
ENSMUST00000177229.8 ENSMUST00000124058.8 |
Foxp1
|
forkhead box P1 |
| chr9_+_66065488 | 0.38 |
ENSMUST00000034944.9
ENSMUST00000238682.2 |
Dapk2
|
death-associated protein kinase 2 |
| chr15_-_5137975 | 0.37 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr8_-_106723026 | 0.37 |
ENSMUST00000227363.2
ENSMUST00000081998.13 |
Dpep2
|
dipeptidase 2 |
| chr17_+_29077385 | 0.36 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr4_+_137720326 | 0.35 |
ENSMUST00000139759.8
ENSMUST00000058133.10 ENSMUST00000105830.9 ENSMUST00000084215.12 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr7_-_30259253 | 0.33 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr17_-_34847354 | 0.32 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_78356523 | 0.31 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr5_-_124325213 | 0.31 |
ENSMUST00000161938.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chrX_-_107877909 | 0.29 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr2_+_24235300 | 0.27 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr2_-_34261121 | 0.27 |
ENSMUST00000127353.3
ENSMUST00000141653.3 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr4_-_82803384 | 0.27 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr1_+_172525613 | 0.27 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr2_+_164339438 | 0.26 |
ENSMUST00000103101.11
ENSMUST00000117066.8 |
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr2_+_36795692 | 0.26 |
ENSMUST00000217479.2
|
Olfr354
|
olfactory receptor 354 |
| chr3_-_102871440 | 0.25 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr10_-_57408585 | 0.25 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr17_-_79292856 | 0.24 |
ENSMUST00000118991.2
|
Prkd3
|
protein kinase D3 |
| chr12_+_55211069 | 0.24 |
ENSMUST00000218889.2
|
Srp54b
|
signal recognition particle 54B |
| chr19_+_26600820 | 0.24 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_112645237 | 0.24 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr4_+_42916666 | 0.23 |
ENSMUST00000132173.8
ENSMUST00000107975.8 |
Phf24
|
PHD finger protein 24 |
| chr12_+_59178258 | 0.23 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr11_+_108811168 | 0.22 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr2_-_70885877 | 0.22 |
ENSMUST00000090849.6
ENSMUST00000100037.9 ENSMUST00000112186.9 |
Mettl8
|
methyltransferase like 8 |
| chr12_+_59178072 | 0.22 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr5_+_123280250 | 0.21 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr15_-_80989200 | 0.21 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr13_+_83672965 | 0.21 |
ENSMUST00000199432.5
ENSMUST00000198069.5 ENSMUST00000197681.5 ENSMUST00000197722.5 ENSMUST00000197938.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr11_-_75918551 | 0.20 |
ENSMUST00000021207.7
|
Rflnb
|
refilin B |
| chr16_+_51851588 | 0.20 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr18_-_60724855 | 0.19 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr2_+_69553141 | 0.19 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr9_-_42855775 | 0.19 |
ENSMUST00000114865.8
|
Grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr2_+_69553384 | 0.19 |
ENSMUST00000040915.15
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr7_+_108209994 | 0.19 |
ENSMUST00000209296.3
|
Olfr506
|
olfactory receptor 506 |
| chr4_-_52859227 | 0.17 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr10_-_128579879 | 0.17 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr17_+_47815998 | 0.17 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr13_+_89687915 | 0.17 |
ENSMUST00000022108.9
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr19_-_37184692 | 0.16 |
ENSMUST00000132580.8
ENSMUST00000079754.11 ENSMUST00000136286.8 ENSMUST00000126188.8 ENSMUST00000126781.2 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr11_+_70323452 | 0.16 |
ENSMUST00000084954.13
ENSMUST00000108568.10 ENSMUST00000079056.9 ENSMUST00000102564.11 ENSMUST00000124943.8 ENSMUST00000150076.8 ENSMUST00000102563.2 |
Arrb2
|
arrestin, beta 2 |
| chr12_-_32000209 | 0.16 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr5_+_102629240 | 0.16 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_-_36782948 | 0.16 |
ENSMUST00000217215.3
|
Olfr353
|
olfactory receptor 353 |
| chr17_+_34457868 | 0.15 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr17_+_17669082 | 0.15 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr5_-_21150470 | 0.14 |
ENSMUST00000197089.2
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr15_-_42540363 | 0.14 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr13_-_36301466 | 0.14 |
ENSMUST00000053265.8
|
Lyrm4
|
LYR motif containing 4 |
| chr12_-_32000169 | 0.14 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr5_+_102629365 | 0.14 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr9_+_18315714 | 0.14 |
ENSMUST00000164441.2
ENSMUST00000169398.3 |
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr8_+_70826191 | 0.14 |
ENSMUST00000003659.9
|
Comp
|
cartilage oligomeric matrix protein |
| chr11_+_4014841 | 0.13 |
ENSMUST00000068322.7
|
Sec14l3
|
SEC14-like lipid binding 3 |
| chr10_+_59942274 | 0.13 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr17_+_33410276 | 0.12 |
ENSMUST00000214406.2
ENSMUST00000213731.2 |
Olfr239
|
olfactory receptor 239 |
| chr6_+_97968737 | 0.12 |
ENSMUST00000043628.13
ENSMUST00000203938.2 |
Mitf
|
melanogenesis associated transcription factor |
| chr14_-_50538979 | 0.12 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr7_+_106692427 | 0.12 |
ENSMUST00000210568.4
|
Olfr17
|
olfactory receptor 17 |
| chr1_-_64160557 | 0.11 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr13_+_23398297 | 0.11 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chr9_-_71070506 | 0.11 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr15_-_99717956 | 0.11 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr14_+_32321341 | 0.11 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr9_+_70586298 | 0.11 |
ENSMUST00000144537.2
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr2_+_164675697 | 0.11 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr2_+_140237229 | 0.11 |
ENSMUST00000110067.8
ENSMUST00000110063.8 ENSMUST00000110064.8 ENSMUST00000110062.8 ENSMUST00000043836.8 ENSMUST00000078027.12 |
Macrod2
|
mono-ADP ribosylhydrolase 2 |
| chr17_+_25097199 | 0.10 |
ENSMUST00000050714.8
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr2_-_51039112 | 0.10 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr6_-_86742789 | 0.10 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4 |
| chr11_-_83177548 | 0.10 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr17_+_37670473 | 0.10 |
ENSMUST00000178766.3
ENSMUST00000215398.2 |
Olfr104-ps
|
olfactory receptor 104, pseudogene |
| chr10_-_128016135 | 0.09 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr1_+_171156568 | 0.09 |
ENSMUST00000111300.8
|
Dedd
|
death effector domain-containing |
| chr3_-_113166153 | 0.09 |
ENSMUST00000098673.5
|
Amy2a5
|
amylase 2a5 |
| chr18_+_38809771 | 0.09 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr6_-_86742847 | 0.09 |
ENSMUST00000113675.8
|
Anxa4
|
annexin A4 |
| chr7_+_30450896 | 0.08 |
ENSMUST00000182229.8
ENSMUST00000080518.14 ENSMUST00000182227.8 ENSMUST00000182721.8 |
Sbsn
|
suprabasin |
| chr2_-_39116284 | 0.08 |
ENSMUST00000204701.3
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr4_+_42922253 | 0.08 |
ENSMUST00000139100.2
|
Phf24
|
PHD finger protein 24 |
| chr2_+_153003212 | 0.08 |
ENSMUST00000089027.3
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr5_-_74189898 | 0.08 |
ENSMUST00000152408.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr18_-_60635059 | 0.08 |
ENSMUST00000042710.8
|
Smim3
|
small integral membrane protein 3 |
| chr14_-_50479323 | 0.08 |
ENSMUST00000214152.2
|
Olfr731
|
olfactory receptor 731 |
| chr8_-_26275182 | 0.08 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr12_-_81014755 | 0.08 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr2_-_163486998 | 0.08 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chrX_-_104919201 | 0.08 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr2_-_39116457 | 0.08 |
ENSMUST00000028087.6
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr3_-_20329823 | 0.07 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr6_-_53955647 | 0.07 |
ENSMUST00000204674.3
ENSMUST00000166545.2 ENSMUST00000203101.3 |
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr9_-_44714263 | 0.07 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr3_+_66892979 | 0.07 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr2_-_104573179 | 0.06 |
ENSMUST00000028595.8
|
Depdc7
|
DEP domain containing 7 |
| chr12_-_100865783 | 0.06 |
ENSMUST00000053668.10
|
Gpr68
|
G protein-coupled receptor 68 |
| chr1_+_157353696 | 0.06 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr19_-_12773472 | 0.06 |
ENSMUST00000038627.9
|
Zfp91
|
zinc finger protein 91 |
| chr3_-_80710097 | 0.06 |
ENSMUST00000075316.10
ENSMUST00000107745.8 |
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| chr13_+_23191826 | 0.06 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr3_+_107137924 | 0.06 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr11_+_78389913 | 0.06 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr12_-_32000534 | 0.05 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr10_+_21868114 | 0.05 |
ENSMUST00000150089.8
ENSMUST00000100036.10 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_+_60878802 | 0.05 |
ENSMUST00000236876.2
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr11_+_46126274 | 0.05 |
ENSMUST00000060185.3
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr8_-_65582206 | 0.05 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr11_-_69088635 | 0.05 |
ENSMUST00000094078.4
ENSMUST00000021262.10 |
Alox8
|
arachidonate 8-lipoxygenase |
| chr17_-_45744637 | 0.05 |
ENSMUST00000024727.10
|
Cdc5l
|
cell division cycle 5-like (S. pombe) |
| chr8_+_4303067 | 0.05 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr9_+_35819708 | 0.04 |
ENSMUST00000176049.2
ENSMUST00000176153.2 |
Pate13
|
prostate and testis expressed 13 |
| chr13_-_96678844 | 0.04 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr12_+_56742413 | 0.04 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9 |
| chr13_+_36301331 | 0.04 |
ENSMUST00000021857.13
|
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr1_+_130793406 | 0.04 |
ENSMUST00000038829.7
|
Fcmr
|
Fc fragment of IgM receptor |
| chr4_+_137720755 | 0.04 |
ENSMUST00000084214.12
ENSMUST00000105831.9 ENSMUST00000203828.3 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr5_-_28672091 | 0.04 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr9_-_101076198 | 0.04 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_-_69696428 | 0.04 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr18_-_39051695 | 0.04 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr16_+_33338648 | 0.04 |
ENSMUST00000122427.8
ENSMUST00000059056.15 |
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr1_+_173093568 | 0.04 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr15_-_54783357 | 0.04 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr8_+_106154266 | 0.04 |
ENSMUST00000067305.8
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr9_+_53757448 | 0.04 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr5_-_103359117 | 0.04 |
ENSMUST00000112846.8
ENSMUST00000170792.9 ENSMUST00000112847.9 ENSMUST00000238446.3 ENSMUST00000133069.8 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr18_-_88945571 | 0.04 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr1_+_75119472 | 0.03 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr13_-_35108293 | 0.03 |
ENSMUST00000223834.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr2_+_96148418 | 0.03 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr11_-_46280281 | 0.03 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr19_+_44980565 | 0.03 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr1_+_93301596 | 0.03 |
ENSMUST00000058682.11
ENSMUST00000186641.7 |
Ano7
|
anoctamin 7 |
| chr10_+_127734384 | 0.03 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr6_-_23132977 | 0.03 |
ENSMUST00000031707.14
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr7_+_113365356 | 0.03 |
ENSMUST00000084696.6
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chrX_-_100266032 | 0.03 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr10_-_95678786 | 0.03 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr4_-_43823866 | 0.03 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr14_-_30665232 | 0.03 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr19_-_9065309 | 0.03 |
ENSMUST00000025554.3
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr15_+_54274151 | 0.03 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr12_+_31315270 | 0.03 |
ENSMUST00000002979.16
ENSMUST00000239496.2 ENSMUST00000170495.3 |
Lamb1
|
laminin B1 |
| chr4_-_102883905 | 0.03 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr12_-_84497718 | 0.03 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr1_-_132318039 | 0.02 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr11_-_69553451 | 0.02 |
ENSMUST00000018905.12
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr3_+_95532282 | 0.02 |
ENSMUST00000058230.13
ENSMUST00000037983.6 |
Ensa
|
endosulfine alpha |
| chr7_-_131918926 | 0.02 |
ENSMUST00000080215.6
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr2_+_138120401 | 0.02 |
ENSMUST00000075410.5
|
Btbd3
|
BTB (POZ) domain containing 3 |
| chr9_+_32135540 | 0.02 |
ENSMUST00000168954.9
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr18_+_37085673 | 0.02 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr5_-_5714196 | 0.02 |
ENSMUST00000196165.5
ENSMUST00000061008.10 ENSMUST00000135252.3 ENSMUST00000054865.13 |
Cfap69
|
cilia and flagella associated protein 69 |
| chr2_+_152404897 | 0.02 |
ENSMUST00000238626.2
|
Gm17416
|
predicted gene, 17416 |
| chr2_-_167032068 | 0.02 |
ENSMUST00000059826.10
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr13_-_22289994 | 0.02 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr7_+_6463510 | 0.02 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chr9_+_53212871 | 0.02 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr11_+_77656414 | 0.02 |
ENSMUST00000164315.2
|
Myo18a
|
myosin XVIIIA |
| chr13_+_117356808 | 0.02 |
ENSMUST00000022242.9
|
Emb
|
embigin |
| chr6_+_29348068 | 0.02 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr3_-_106733603 | 0.02 |
ENSMUST00000213616.3
|
Olfr266
|
olfactory receptor 266 |
| chr3_-_27764522 | 0.02 |
ENSMUST00000195008.6
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr8_+_94537910 | 0.02 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr7_+_113365235 | 0.02 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr14_+_32321824 | 0.02 |
ENSMUST00000068938.7
ENSMUST00000228878.2 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr9_+_70586232 | 0.02 |
ENSMUST00000067880.13
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr17_-_31383976 | 0.02 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr9_+_110673565 | 0.01 |
ENSMUST00000176403.8
|
Prss46
|
protease, serine 46 |
| chrX_-_140508177 | 0.01 |
ENSMUST00000067841.8
|
Irs4
|
insulin receptor substrate 4 |
| chr1_-_160079007 | 0.01 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr14_-_110992533 | 0.01 |
ENSMUST00000078386.4
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr14_-_50519728 | 0.01 |
ENSMUST00000071208.3
|
Olfr732
|
olfactory receptor 732 |
| chr18_+_52598748 | 0.01 |
ENSMUST00000025406.9
|
Srfbp1
|
serum response factor binding protein 1 |
| chr12_-_81014849 | 0.01 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr14_-_52610508 | 0.01 |
ENSMUST00000071221.2
|
Olfr1512
|
olfactory receptor 1512 |
| chr7_-_103492361 | 0.01 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr7_-_103261741 | 0.01 |
ENSMUST00000052152.3
|
Olfr620
|
olfactory receptor 620 |
| chr7_+_18962301 | 0.01 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chr12_+_8027640 | 0.01 |
ENSMUST00000171271.8
ENSMUST00000037811.13 |
Apob
|
apolipoprotein B |
| chr5_+_88032867 | 0.01 |
ENSMUST00000129757.9
|
Odam
|
odontogenic, ameloblast asssociated |
| chr11_-_99433984 | 0.01 |
ENSMUST00000107443.8
ENSMUST00000074253.4 |
Krt40
|
keratin 40 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.5 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.1 | 0.4 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.3 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.2 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 1.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.4 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.0 | GO:2000729 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 1.6 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |