Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
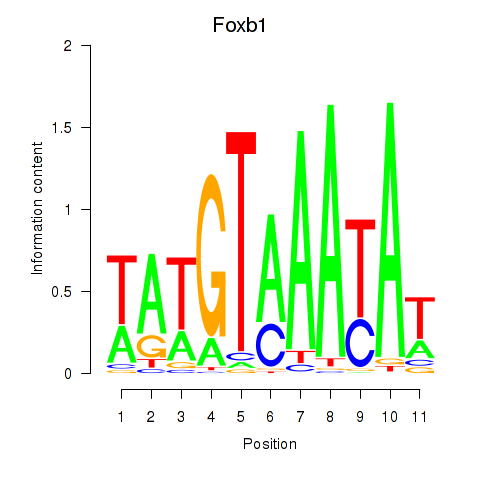
Results for Foxb1
Z-value: 0.55
Transcription factors associated with Foxb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxb1
|
ENSMUSG00000059246.5 | forkhead box B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxb1 | mm39_v1_chr9_-_69668204_69668222 | -0.28 | 6.5e-01 | Click! |
Activity profile of Foxb1 motif
Sorted Z-values of Foxb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_78356523 | 0.42 |
ENSMUST00000001126.4
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr9_-_103569984 | 0.25 |
ENSMUST00000049452.15
|
Tmem108
|
transmembrane protein 108 |
| chr5_+_102629240 | 0.19 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_102629365 | 0.18 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_16758777 | 0.14 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor |
| chr5_-_3707166 | 0.14 |
ENSMUST00000196304.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr13_+_94954202 | 0.14 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr5_+_16758897 | 0.14 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor |
| chr15_+_43340609 | 0.13 |
ENSMUST00000022962.8
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr1_+_43484895 | 0.12 |
ENSMUST00000086421.9
|
Nck2
|
non-catalytic region of tyrosine kinase adaptor protein 2 |
| chr12_-_32000169 | 0.12 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr5_+_16758538 | 0.12 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor |
| chr18_-_25886750 | 0.11 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr14_-_55880708 | 0.10 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr7_+_119499322 | 0.10 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr2_+_29951859 | 0.10 |
ENSMUST00000102866.10
|
Set
|
SET nuclear oncogene |
| chr1_+_179788037 | 0.09 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_-_96147592 | 0.09 |
ENSMUST00000074976.8
|
H2ac19
|
H2A clustered histone 19 |
| chr2_+_130119077 | 0.08 |
ENSMUST00000028890.15
ENSMUST00000159373.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr7_+_120442048 | 0.08 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr3_-_146487102 | 0.08 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chrX_+_108138965 | 0.08 |
ENSMUST00000033598.9
|
Sh3bgrl
|
SH3-binding domain glutamic acid-rich protein like |
| chr7_+_27290969 | 0.08 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr7_-_99994257 | 0.07 |
ENSMUST00000207634.2
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr3_+_96152813 | 0.07 |
ENSMUST00000078756.7
ENSMUST00000090779.4 |
H2ac18
Gm20634
|
H2A clustered histone 18 predicted gene 20634 |
| chr17_+_8144822 | 0.07 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr3_+_85946145 | 0.07 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr12_+_112645237 | 0.07 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr9_-_32452885 | 0.07 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr2_-_45001141 | 0.06 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_27291126 | 0.06 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr8_+_108271643 | 0.06 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_179788675 | 0.06 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_-_97904958 | 0.05 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr17_+_35188888 | 0.05 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481 |
| chr4_-_87724533 | 0.05 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr2_+_29951766 | 0.05 |
ENSMUST00000149578.8
|
Set
|
SET nuclear oncogene |
| chr10_-_14420725 | 0.04 |
ENSMUST00000041168.6
ENSMUST00000238680.2 |
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr6_+_14901343 | 0.04 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr6_+_34840057 | 0.04 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr16_-_33916354 | 0.04 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr17_-_71158703 | 0.03 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr7_+_67305162 | 0.03 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr2_-_86906161 | 0.03 |
ENSMUST00000214049.2
|
Olfr1107
|
olfactory receptor 1107 |
| chr5_+_66833434 | 0.03 |
ENSMUST00000031131.11
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr10_+_38841511 | 0.03 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr17_+_48037758 | 0.03 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chrX_-_93166693 | 0.03 |
ENSMUST00000113925.8
|
Zfx
|
zinc finger protein X-linked |
| chr8_-_62576140 | 0.03 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr16_-_74208180 | 0.03 |
ENSMUST00000117200.8
|
Robo2
|
roundabout guidance receptor 2 |
| chr15_-_55411560 | 0.03 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr2_-_163486998 | 0.03 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr17_+_12597490 | 0.02 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr10_+_69761597 | 0.02 |
ENSMUST00000182269.8
ENSMUST00000183261.8 ENSMUST00000183074.8 |
Ank3
|
ankyrin 3, epithelial |
| chr16_+_44215136 | 0.02 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr10_-_61814852 | 0.02 |
ENSMUST00000105453.8
ENSMUST00000105452.9 ENSMUST00000105454.3 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr19_+_5091365 | 0.02 |
ENSMUST00000116567.4
ENSMUST00000235918.2 |
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr18_+_37085673 | 0.02 |
ENSMUST00000192512.6
ENSMUST00000192295.2 ENSMUST00000115661.5 |
Pcdha4
Gm42416
|
protocadherin alpha 4 predicted gene, 42416 |
| chr16_-_44153498 | 0.02 |
ENSMUST00000047446.13
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr10_+_69761784 | 0.02 |
ENSMUST00000181974.8
ENSMUST00000182795.8 ENSMUST00000182437.8 |
Ank3
|
ankyrin 3, epithelial |
| chr8_+_94537910 | 0.02 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr19_+_47217279 | 0.02 |
ENSMUST00000111807.5
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr2_-_27365633 | 0.02 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr8_-_45747883 | 0.02 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr8_+_106785434 | 0.02 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr1_+_70764874 | 0.02 |
ENSMUST00000053922.12
ENSMUST00000161937.2 ENSMUST00000162182.2 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr3_+_55689921 | 0.02 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr6_+_132844971 | 0.02 |
ENSMUST00000082014.2
|
Tas2r110
|
taste receptor, type 2, member 110 |
| chrX_+_56257374 | 0.02 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr9_+_53757448 | 0.02 |
ENSMUST00000048485.7
|
Sln
|
sarcolipin |
| chr14_+_33193765 | 0.02 |
ENSMUST00000208577.2
|
Frmpd2
|
FERM and PDZ domain containing 2 |
| chr4_-_102883905 | 0.02 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr17_-_21110913 | 0.02 |
ENSMUST00000061278.2
|
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chr7_-_119078472 | 0.02 |
ENSMUST00000209095.2
ENSMUST00000033263.6 ENSMUST00000207261.2 |
Umod
|
uromodulin |
| chr15_-_48655329 | 0.02 |
ENSMUST00000160658.8
ENSMUST00000100670.10 ENSMUST00000162830.8 |
Csmd3
|
CUB and Sushi multiple domains 3 |
| chr10_+_69761314 | 0.01 |
ENSMUST00000182692.8
ENSMUST00000092433.12 |
Ank3
|
ankyrin 3, epithelial |
| chr2_-_134396268 | 0.01 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr4_+_32238950 | 0.01 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr2_-_147028309 | 0.01 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr8_-_45715049 | 0.01 |
ENSMUST00000034064.5
|
F11
|
coagulation factor XI |
| chr3_+_133942244 | 0.01 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr1_+_187995096 | 0.01 |
ENSMUST00000060479.14
|
Ush2a
|
usherin |
| chr6_-_3494587 | 0.01 |
ENSMUST00000049985.15
|
Hepacam2
|
HEPACAM family member 2 |
| chr17_+_70829050 | 0.01 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr16_-_44153288 | 0.01 |
ENSMUST00000136381.8
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_+_109092610 | 0.01 |
ENSMUST00000106628.8
|
Calr4
|
calreticulin 4 |
| chr11_-_99328969 | 0.01 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr3_-_113325938 | 0.01 |
ENSMUST00000132353.2
|
Amy2a1
|
amylase 2a1 |
| chr12_-_32000209 | 0.01 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr18_-_43820759 | 0.01 |
ENSMUST00000082254.8
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr3_+_75982890 | 0.01 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr19_-_38031774 | 0.01 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr7_-_37472979 | 0.01 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr5_+_104318542 | 0.01 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr11_-_16958647 | 0.01 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr19_-_40175709 | 0.01 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr19_-_9065309 | 0.01 |
ENSMUST00000025554.3
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin) |
| chr5_+_43673093 | 0.01 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr17_+_93506590 | 0.01 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr1_-_158642039 | 0.01 |
ENSMUST00000161589.3
|
Pappa2
|
pappalysin 2 |
| chr3_-_123483772 | 0.01 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr3_-_33136153 | 0.01 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr17_+_93506435 | 0.01 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr7_-_106473450 | 0.01 |
ENSMUST00000060879.6
|
Olfr705
|
olfactory receptor 705 |
| chr8_-_65489834 | 0.01 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr10_+_69761630 | 0.01 |
ENSMUST00000182029.8
|
Ank3
|
ankyrin 3, epithelial |
| chr7_-_144761806 | 0.01 |
ENSMUST00000208788.2
|
Smim38
|
small integral membrane protein 38 |
| chr4_-_82970384 | 0.01 |
ENSMUST00000170248.9
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr2_+_154042291 | 0.01 |
ENSMUST00000028987.7
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr18_-_47466378 | 0.01 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrX_+_162923474 | 0.01 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr5_-_87630117 | 0.01 |
ENSMUST00000079811.13
ENSMUST00000144144.3 |
Ugt2a2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr2_+_109522781 | 0.01 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr2_+_86616076 | 0.01 |
ENSMUST00000055273.2
|
Olfr1093
|
olfactory receptor 1093 |
| chr2_+_181408833 | 0.01 |
ENSMUST00000108756.8
|
Myt1
|
myelin transcription factor 1 |
| chr12_+_119291343 | 0.01 |
ENSMUST00000221917.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chrX_+_100419965 | 0.01 |
ENSMUST00000119080.8
|
Gjb1
|
gap junction protein, beta 1 |
| chr6_-_139478919 | 0.01 |
ENSMUST00000170650.3
|
Rergl
|
RERG/RAS-like |
| chr10_-_33662700 | 0.01 |
ENSMUST00000223295.2
|
Sult3a2
|
sulfotransferase family 3A, member 2 |
| chr3_-_92922976 | 0.01 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr1_+_81054921 | 0.01 |
ENSMUST00000123720.8
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr10_+_129357661 | 0.01 |
ENSMUST00000217228.2
|
Olfr791
|
olfactory receptor 791 |
| chr6_-_41535292 | 0.01 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr6_+_34840151 | 0.01 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr1_+_19279178 | 0.01 |
ENSMUST00000187754.7
|
Tfap2b
|
transcription factor AP-2 beta |
| chr17_+_70829144 | 0.01 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr13_+_21316868 | 0.01 |
ENSMUST00000096006.3
|
Olfr263
|
olfactory receptor 263 |
| chr11_-_99877423 | 0.01 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr6_-_41535322 | 0.01 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr5_-_51725059 | 0.01 |
ENSMUST00000127135.3
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| chr19_+_20579322 | 0.01 |
ENSMUST00000087638.4
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1 |
| chr8_+_66838927 | 0.01 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr3_-_95811993 | 0.01 |
ENSMUST00000147962.3
ENSMUST00000036181.15 |
Car14
|
carbonic anhydrase 14 |
| chr19_-_20704896 | 0.01 |
ENSMUST00000025656.4
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr4_+_109092829 | 0.01 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr4_-_139560229 | 0.01 |
ENSMUST00000174681.2
|
Pax7
|
paired box 7 |
| chr10_+_127734384 | 0.01 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_-_103492361 | 0.01 |
ENSMUST00000063957.6
|
Hbb-bh1
|
hemoglobin Z, beta-like embryonic chain |
| chr5_+_104473195 | 0.01 |
ENSMUST00000066207.4
|
Mepe
|
matrix extracellular phosphoglycoprotein with ASARM motif (bone) |
| chr7_-_115459082 | 0.01 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr4_+_127066667 | 0.01 |
ENSMUST00000106094.9
|
Dlgap3
|
DLG associated protein 3 |
| chr6_+_8520006 | 0.01 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr2_-_60552980 | 0.01 |
ENSMUST00000028348.9
ENSMUST00000112517.8 |
Itgb6
|
integrin beta 6 |
| chr2_+_181409075 | 0.01 |
ENSMUST00000108757.9
|
Myt1
|
myelin transcription factor 1 |
| chr3_-_129597679 | 0.00 |
ENSMUST00000185462.7
ENSMUST00000179187.2 |
Lrit3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr2_-_86982968 | 0.00 |
ENSMUST00000216378.2
|
Olfr1111
|
olfactory receptor 1111 |
| chrX_-_42363663 | 0.00 |
ENSMUST00000016294.8
|
Tenm1
|
teneurin transmembrane protein 1 |
| chr12_-_40249314 | 0.00 |
ENSMUST00000095760.3
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr4_+_102287244 | 0.00 |
ENSMUST00000172616.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_+_7494108 | 0.00 |
ENSMUST00000193330.2
|
Pkia
|
protein kinase inhibitor, alpha |
| chr6_+_5390386 | 0.00 |
ENSMUST00000183358.2
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr12_-_111946560 | 0.00 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr3_-_146302343 | 0.00 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr4_-_139560831 | 0.00 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr6_-_23248361 | 0.00 |
ENSMUST00000031709.7
|
Fezf1
|
Fez family zinc finger 1 |
| chr10_-_49659355 | 0.00 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr17_-_3746536 | 0.00 |
ENSMUST00000115800.2
|
Nox3
|
NADPH oxidase 3 |
| chr7_-_141009264 | 0.00 |
ENSMUST00000164387.2
ENSMUST00000137488.2 ENSMUST00000084436.10 |
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chr12_+_56742413 | 0.00 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9 |
| chr17_-_35265702 | 0.00 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr11_-_98220466 | 0.00 |
ENSMUST00000041685.7
|
Neurod2
|
neurogenic differentiation 2 |
| chr6_+_8948608 | 0.00 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
| chr7_-_141009346 | 0.00 |
ENSMUST00000124444.2
|
Cend1
|
cell cycle exit and neuronal differentiation 1 |
| chrY_-_797405 | 0.00 |
ENSMUST00000189888.7
ENSMUST00000065545.6 |
Zfy1
|
zinc finger protein 1, Y-linked |
| chr6_+_14901439 | 0.00 |
ENSMUST00000128567.8
|
Foxp2
|
forkhead box P2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) methotrexate transport(GO:0051958) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |