Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
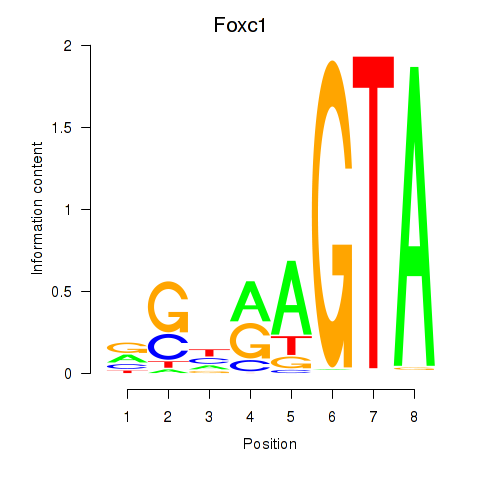
Results for Foxc1
Z-value: 0.41
Transcription factors associated with Foxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc1
|
ENSMUSG00000050295.5 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc1 | mm39_v1_chr13_+_31990604_31990633 | -0.88 | 4.9e-02 | Click! |
Activity profile of Foxc1 motif
Sorted Z-values of Foxc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_78392969 | 0.18 |
ENSMUST00000164064.2
|
Gm6133
|
predicted gene 6133 |
| chr9_-_103099262 | 0.17 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr14_-_31362835 | 0.17 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr11_-_78441584 | 0.17 |
ENSMUST00000103242.5
|
Tmem97
|
transmembrane protein 97 |
| chr3_+_37366662 | 0.16 |
ENSMUST00000138710.3
ENSMUST00000057975.8 ENSMUST00000108121.4 |
Gm43439
Bbs12
|
predicted gene 43439 Bardet-Biedl syndrome 12 (human) |
| chr1_-_131025562 | 0.16 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr3_+_68598757 | 0.15 |
ENSMUST00000107816.4
|
Il12a
|
interleukin 12a |
| chr7_+_119360141 | 0.14 |
ENSMUST00000106528.8
ENSMUST00000106527.8 ENSMUST00000063770.10 ENSMUST00000106529.8 |
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr2_+_30697838 | 0.14 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr15_-_78290038 | 0.14 |
ENSMUST00000058659.9
|
Tst
|
thiosulfate sulfurtransferase, mitochondrial |
| chr15_+_76784110 | 0.14 |
ENSMUST00000068407.6
ENSMUST00000109793.3 |
Commd5
|
COMM domain containing 5 |
| chr9_-_106768601 | 0.14 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr9_+_120762466 | 0.13 |
ENSMUST00000007130.15
ENSMUST00000178812.9 |
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr6_-_124410452 | 0.13 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr4_-_116484675 | 0.13 |
ENSMUST00000081182.5
ENSMUST00000030457.12 |
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr10_+_39245746 | 0.13 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr8_-_46605196 | 0.12 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr10_+_127157784 | 0.12 |
ENSMUST00000219511.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr6_-_83031358 | 0.12 |
ENSMUST00000113962.8
ENSMUST00000089645.13 ENSMUST00000113963.8 |
Htra2
|
HtrA serine peptidase 2 |
| chr1_+_75213044 | 0.12 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr6_-_67512768 | 0.12 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr3_-_89905547 | 0.12 |
ENSMUST00000199740.2
ENSMUST00000198782.2 |
Hax1
|
HCLS1 associated X-1 |
| chr11_-_96720309 | 0.12 |
ENSMUST00000167149.8
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr11_-_74615496 | 0.11 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr8_+_84379298 | 0.11 |
ENSMUST00000019577.10
ENSMUST00000211985.2 ENSMUST00000212463.2 |
Gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr7_-_110682204 | 0.11 |
ENSMUST00000161051.8
ENSMUST00000160132.8 ENSMUST00000162415.9 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr12_-_101943134 | 0.11 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr13_-_8921732 | 0.11 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr11_+_49327451 | 0.10 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr1_-_16689660 | 0.10 |
ENSMUST00000117146.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr9_+_7558449 | 0.10 |
ENSMUST00000018765.4
|
Mmp8
|
matrix metallopeptidase 8 |
| chr10_+_127159609 | 0.10 |
ENSMUST00000069548.7
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr1_+_163822438 | 0.10 |
ENSMUST00000045694.14
ENSMUST00000111490.2 |
Mettl18
|
methyltransferase like 18 |
| chr7_+_79048884 | 0.10 |
ENSMUST00000137667.3
|
Fanci
|
Fanconi anemia, complementation group I |
| chr5_-_114961501 | 0.10 |
ENSMUST00000100850.6
|
1500011B03Rik
|
RIKEN cDNA 1500011B03 gene |
| chrX_-_163033364 | 0.09 |
ENSMUST00000112263.2
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr2_-_181007099 | 0.09 |
ENSMUST00000108808.8
ENSMUST00000170190.8 ENSMUST00000127988.8 |
Arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr5_-_149559636 | 0.09 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr7_+_127503812 | 0.09 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr12_-_65010124 | 0.09 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr2_+_163836880 | 0.09 |
ENSMUST00000018470.10
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr12_-_73093953 | 0.09 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr14_-_31362909 | 0.09 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr8_-_46604742 | 0.09 |
ENSMUST00000041582.15
|
Snx25
|
sorting nexin 25 |
| chr11_+_100973391 | 0.09 |
ENSMUST00000001806.10
ENSMUST00000107308.4 |
Coasy
|
Coenzyme A synthase |
| chr4_-_116485118 | 0.09 |
ENSMUST00000030456.14
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr5_-_149559667 | 0.09 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr14_+_113552034 | 0.09 |
ENSMUST00000072359.8
|
Tpm3-rs7
|
tropomyosin 3, related sequence 7 |
| chr3_+_40904253 | 0.08 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chrY_+_1010543 | 0.08 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr11_-_78642480 | 0.08 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr4_+_97665843 | 0.08 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr7_-_5128936 | 0.08 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr9_-_96513529 | 0.08 |
ENSMUST00000034984.8
|
Rasa2
|
RAS p21 protein activator 2 |
| chr9_+_108447077 | 0.07 |
ENSMUST00000019183.14
|
Dalrd3
|
DALR anticodon binding domain containing 3 |
| chr16_+_35803794 | 0.07 |
ENSMUST00000173555.8
|
Kpna1
|
karyopherin (importin) alpha 1 |
| chr8_-_78337297 | 0.07 |
ENSMUST00000141202.2
ENSMUST00000152168.8 |
Tmem184c
|
transmembrane protein 184C |
| chr14_+_31363004 | 0.07 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chrX_-_108056995 | 0.07 |
ENSMUST00000033597.9
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr1_-_16689527 | 0.07 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr10_+_127159568 | 0.06 |
ENSMUST00000219026.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr4_+_117407517 | 0.06 |
ENSMUST00000037127.15
|
Eri3
|
exoribonuclease 3 |
| chr2_+_181007177 | 0.06 |
ENSMUST00000108807.9
|
Zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr1_+_75213114 | 0.06 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr7_+_18962252 | 0.06 |
ENSMUST00000063976.9
|
Opa3
|
optic atrophy 3 |
| chr5_-_52827015 | 0.06 |
ENSMUST00000031069.13
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr11_-_3881789 | 0.06 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr10_-_14581203 | 0.06 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr9_-_89586960 | 0.06 |
ENSMUST00000058488.9
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr1_+_134110142 | 0.06 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr1_+_75213082 | 0.05 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr14_-_52252432 | 0.05 |
ENSMUST00000226527.2
|
Zfp219
|
zinc finger protein 219 |
| chr15_-_57897702 | 0.05 |
ENSMUST00000100655.5
ENSMUST00000185553.2 |
9130401M01Rik
|
RIKEN cDNA 9130401M01 gene |
| chr11_-_4544751 | 0.05 |
ENSMUST00000109943.10
|
Mtmr3
|
myotubularin related protein 3 |
| chr19_-_24202344 | 0.05 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr11_-_71092124 | 0.05 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr15_-_10485976 | 0.05 |
ENSMUST00000169050.8
ENSMUST00000022855.12 |
Brix1
|
BRX1, biogenesis of ribosomes |
| chr16_-_59452883 | 0.05 |
ENSMUST00000118438.8
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr3_-_89905927 | 0.04 |
ENSMUST00000197725.5
ENSMUST00000197767.5 ENSMUST00000197786.5 ENSMUST00000079724.9 |
Hax1
|
HCLS1 associated X-1 |
| chr15_+_6451721 | 0.04 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr9_+_110248815 | 0.04 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr2_+_127050281 | 0.04 |
ENSMUST00000103220.4
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr1_+_75213171 | 0.04 |
ENSMUST00000187058.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr3_-_37180093 | 0.04 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr4_-_49521036 | 0.04 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr3_-_138780894 | 0.04 |
ENSMUST00000196280.5
ENSMUST00000200396.2 |
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chr19_-_53932581 | 0.04 |
ENSMUST00000236885.2
ENSMUST00000236098.2 ENSMUST00000236370.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr17_-_33904386 | 0.04 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr17_-_33904345 | 0.04 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr5_+_8096074 | 0.04 |
ENSMUST00000088786.11
|
Sri
|
sorcin |
| chr7_-_89590230 | 0.04 |
ENSMUST00000075010.12
|
Hikeshi
|
heat shock protein nuclear import factor |
| chr2_+_62494622 | 0.04 |
ENSMUST00000028257.3
|
Gca
|
grancalcin |
| chr15_+_78481247 | 0.04 |
ENSMUST00000043069.6
ENSMUST00000231180.2 ENSMUST00000229796.2 ENSMUST00000229295.2 |
Cyth4
|
cytohesin 4 |
| chr9_+_54771064 | 0.04 |
ENSMUST00000034843.9
|
Ireb2
|
iron responsive element binding protein 2 |
| chr19_-_53932867 | 0.04 |
ENSMUST00000235688.2
ENSMUST00000235348.2 |
Bbip1
|
BBSome interacting protein 1 |
| chr4_-_59783780 | 0.04 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr14_-_52252318 | 0.04 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_-_96104886 | 0.04 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr11_-_71092282 | 0.03 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chrX_-_74460168 | 0.03 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr1_+_45350698 | 0.03 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr1_-_4430481 | 0.03 |
ENSMUST00000027032.6
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chr16_-_43709968 | 0.03 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr8_+_46604786 | 0.03 |
ENSMUST00000154040.2
|
Cfap97
|
cilia and flagella associated protein 97 |
| chr1_-_168259839 | 0.03 |
ENSMUST00000188912.7
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr9_+_50686647 | 0.03 |
ENSMUST00000159576.2
|
Alg9
|
asparagine-linked glycosylation 9 (alpha 1,2 mannosyltransferase) |
| chrX_+_65696608 | 0.03 |
ENSMUST00000036043.5
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr13_-_74918701 | 0.03 |
ENSMUST00000223126.2
|
Cast
|
calpastatin |
| chr2_+_109522781 | 0.03 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr7_+_141996067 | 0.03 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr6_-_52190299 | 0.03 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr5_-_135601887 | 0.03 |
ENSMUST00000004936.10
ENSMUST00000201401.2 |
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chr9_+_44245981 | 0.03 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr5_-_100186728 | 0.03 |
ENSMUST00000153442.8
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr8_-_84769170 | 0.03 |
ENSMUST00000005601.9
|
Il27ra
|
interleukin 27 receptor, alpha |
| chrX_-_56384089 | 0.02 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr12_+_76417040 | 0.02 |
ENSMUST00000042779.4
|
Zbtb1
|
zinc finger and BTB domain containing 1 |
| chr10_+_81464370 | 0.02 |
ENSMUST00000123896.8
ENSMUST00000153573.2 ENSMUST00000119336.8 |
Ankrd24
|
ankyrin repeat domain 24 |
| chr3_+_68375495 | 0.02 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr1_+_131890679 | 0.02 |
ENSMUST00000191034.2
ENSMUST00000177943.8 |
Gm29103
Slc45a3
|
predicted gene 29103 solute carrier family 45, member 3 |
| chr3_+_89986925 | 0.02 |
ENSMUST00000118566.8
ENSMUST00000119158.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr10_+_45453907 | 0.02 |
ENSMUST00000037044.13
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr2_-_172212426 | 0.02 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr16_-_19703014 | 0.02 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chr1_-_168259710 | 0.02 |
ENSMUST00000072863.6
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr6_-_129637519 | 0.02 |
ENSMUST00000119533.2
ENSMUST00000145984.8 ENSMUST00000118401.8 ENSMUST00000112057.9 ENSMUST00000071920.11 |
Klrc2
|
killer cell lectin-like receptor subfamily C, member 2 |
| chr6_+_41118120 | 0.02 |
ENSMUST00000103273.3
|
Trbv15
|
T cell receptor beta, variable 15 |
| chr16_+_41353360 | 0.02 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr15_+_80507671 | 0.02 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr19_+_29229826 | 0.02 |
ENSMUST00000238009.2
|
Jak2
|
Janus kinase 2 |
| chr19_-_11301919 | 0.02 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr10_-_129781929 | 0.02 |
ENSMUST00000074308.3
|
Olfr818
|
olfactory receptor 818 |
| chr3_-_87170903 | 0.02 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr9_-_71499628 | 0.02 |
ENSMUST00000093823.8
|
Myzap
|
myocardial zonula adherens protein |
| chr14_-_99337137 | 0.02 |
ENSMUST00000042471.11
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr12_-_80807454 | 0.02 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr18_+_37948443 | 0.02 |
ENSMUST00000195239.2
|
Pcdhgc4
|
protocadherin gamma subfamily C, 4 |
| chr11_+_3152683 | 0.02 |
ENSMUST00000125637.8
|
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr13_-_21330037 | 0.02 |
ENSMUST00000216039.3
|
Olfr1368
|
olfactory receptor 1368 |
| chr14_-_50425655 | 0.02 |
ENSMUST00000205837.3
|
Olfr730
|
olfactory receptor 730 |
| chr7_-_43885552 | 0.02 |
ENSMUST00000236952.2
|
1700028J19Rik
|
RIKEN cDNA 1700028J19 gene |
| chr4_-_94817025 | 0.02 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr6_-_30680502 | 0.02 |
ENSMUST00000133373.8
|
Cep41
|
centrosomal protein 41 |
| chr8_-_120301883 | 0.01 |
ENSMUST00000212198.2
ENSMUST00000212065.2 ENSMUST00000036049.6 |
Hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr9_+_38738911 | 0.01 |
ENSMUST00000051238.7
ENSMUST00000219798.2 |
Olfr923
|
olfactory receptor 923 |
| chr17_+_34457868 | 0.01 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr2_-_85992176 | 0.01 |
ENSMUST00000099908.6
ENSMUST00000215624.2 |
Olfr1042
|
olfactory receptor 1042 |
| chr16_+_37360245 | 0.01 |
ENSMUST00000023524.13
|
Rabl3
|
RAB, member RAS oncogene family-like 3 |
| chr5_-_44259293 | 0.01 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr17_+_48666919 | 0.01 |
ENSMUST00000224001.2
ENSMUST00000024792.8 ENSMUST00000225849.2 |
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr7_-_10347586 | 0.01 |
ENSMUST00000226190.2
ENSMUST00000226228.2 ENSMUST00000228296.2 ENSMUST00000228638.2 ENSMUST00000176707.3 ENSMUST00000176284.9 ENSMUST00000226160.2 ENSMUST00000227853.2 ENSMUST00000228478.2 |
Vmn1r69
|
vomeronasal 1 receptor 69 |
| chr18_-_3281089 | 0.01 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr10_-_23977810 | 0.01 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr17_-_37574640 | 0.01 |
ENSMUST00000080759.5
|
Olfr98
|
olfactory receptor 98 |
| chr9_-_44920899 | 0.01 |
ENSMUST00000102832.3
|
Cd3e
|
CD3 antigen, epsilon polypeptide |
| chr18_+_37884653 | 0.01 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr13_+_19362068 | 0.01 |
ENSMUST00000103553.3
|
Trgv7
|
T cell receptor gamma, variable 7 |
| chr17_+_37356872 | 0.01 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr9_-_51240201 | 0.01 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr15_-_100497863 | 0.01 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr6_+_40763875 | 0.01 |
ENSMUST00000195870.3
|
Mgam2-ps
|
maltase-glucoamylase 2, pseudogene |
| chr4_-_94817056 | 0.01 |
ENSMUST00000107097.9
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr7_+_86234485 | 0.01 |
ENSMUST00000078447.2
|
Olfr295
|
olfactory receptor 295 |
| chr10_-_129010923 | 0.01 |
ENSMUST00000169800.4
|
Olfr772
|
olfactory receptor 772 |
| chr3_+_103483397 | 0.01 |
ENSMUST00000183637.8
ENSMUST00000117221.9 ENSMUST00000118117.8 ENSMUST00000118563.3 |
Syt6
|
synaptotagmin VI |
| chr7_+_30193047 | 0.01 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_24679154 | 0.01 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr6_-_57821483 | 0.01 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr2_-_86705146 | 0.01 |
ENSMUST00000130722.3
|
Olfr1096
|
olfactory receptor 1096 |
| chr1_+_107439145 | 0.01 |
ENSMUST00000009356.11
ENSMUST00000064916.9 |
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr2_+_110551927 | 0.01 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr3_+_94284739 | 0.01 |
ENSMUST00000197040.5
|
Rorc
|
RAR-related orphan receptor gamma |
| chr19_+_40078132 | 0.01 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr14_+_33041660 | 0.01 |
ENSMUST00000111955.2
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr16_+_41353212 | 0.01 |
ENSMUST00000078873.11
|
Lsamp
|
limbic system-associated membrane protein |
| chr10_-_128974829 | 0.01 |
ENSMUST00000203887.3
ENSMUST00000204250.3 ENSMUST00000204712.4 |
Olfr770
|
olfactory receptor 770 |
| chr7_-_85962916 | 0.01 |
ENSMUST00000174158.3
|
Olfr309
|
olfactory receptor 309 |
| chr17_-_24908874 | 0.01 |
ENSMUST00000007236.5
|
Syngr3
|
synaptogyrin 3 |
| chr1_+_135980488 | 0.01 |
ENSMUST00000160641.8
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr3_-_122828592 | 0.01 |
ENSMUST00000029761.14
|
Myoz2
|
myozenin 2 |
| chr13_+_24118417 | 0.01 |
ENSMUST00000072391.2
|
H2ac1
|
H2A clustered histone 1 |
| chr14_-_48900192 | 0.01 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr14_+_53980561 | 0.01 |
ENSMUST00000103667.6
|
Trav16
|
T cell receptor alpha variable 16 |
| chr17_+_37777099 | 0.01 |
ENSMUST00000031086.5
|
Olfr109
|
olfactory receptor 109 |
| chr17_+_37756371 | 0.01 |
ENSMUST00000078207.4
ENSMUST00000218675.2 |
Olfr108
|
olfactory receptor 108 |
| chr16_-_48592319 | 0.01 |
ENSMUST00000239408.2
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr18_-_3280999 | 0.01 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr11_-_58346806 | 0.01 |
ENSMUST00000055204.6
|
Olfr30
|
olfactory receptor 30 |
| chr1_+_135980508 | 0.01 |
ENSMUST00000112068.10
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr1_+_173030925 | 0.01 |
ENSMUST00000057548.2
|
Olfr218
|
olfactory receptor 218 |
| chr2_-_87955337 | 0.01 |
ENSMUST00000099833.6
|
Olfr1166
|
olfactory receptor 1166 |
| chr10_-_116732813 | 0.01 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr14_+_63235512 | 0.01 |
ENSMUST00000100492.5
|
Defb47
|
defensin beta 47 |
| chr2_-_79738734 | 0.01 |
ENSMUST00000090756.11
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr19_+_13478481 | 0.01 |
ENSMUST00000214274.2
|
Olfr1477
|
olfactory receptor 1477 |
| chr10_+_115405891 | 0.01 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr14_+_53913598 | 0.01 |
ENSMUST00000103662.6
|
Trav9-4
|
T cell receptor alpha variable 9-4 |
| chr2_-_79738773 | 0.01 |
ENSMUST00000102652.10
ENSMUST00000102651.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_88022172 | 0.01 |
ENSMUST00000203674.3
ENSMUST00000204126.2 ENSMUST00000113596.8 ENSMUST00000113600.10 |
Rab7
|
RAB7, member RAS oncogene family |
| chr2_+_85600147 | 0.01 |
ENSMUST00000065626.3
|
Olfr1013
|
olfactory receptor 1013 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.1 | GO:1904501 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.2 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.3 | GO:0051135 | positive regulation of NK T cell activation(GO:0051135) |
| 0.0 | 0.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.0 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |