Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
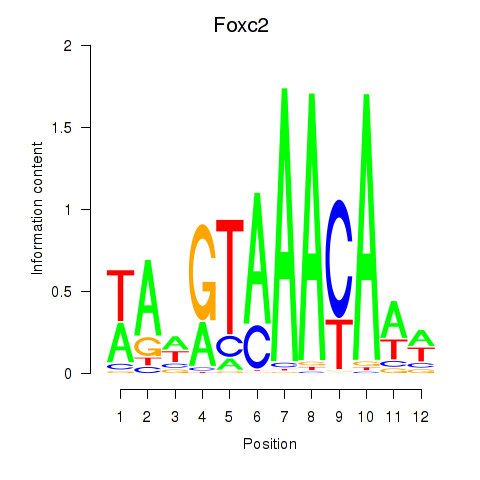
Results for Foxc2
Z-value: 0.53
Transcription factors associated with Foxc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxc2
|
ENSMUSG00000046714.8 | forkhead box C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxc2 | mm39_v1_chr8_+_121842902_121842916 | -0.69 | 2.0e-01 | Click! |
Activity profile of Foxc2 motif
Sorted Z-values of Foxc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_87724533 | 0.30 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr14_+_48358267 | 0.25 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr6_-_137548004 | 0.24 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr8_+_106785434 | 0.24 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr6_+_146934082 | 0.22 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr14_+_71011744 | 0.22 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr1_+_40554513 | 0.21 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr1_+_177272215 | 0.19 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr16_-_43484494 | 0.18 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr19_+_45433899 | 0.18 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr14_+_61844899 | 0.16 |
ENSMUST00000225582.2
ENSMUST00000051184.10 |
Kcnrg
|
potassium channel regulator |
| chr17_+_29309942 | 0.16 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chrX_-_159518743 | 0.15 |
ENSMUST00000135856.2
|
Ppef1
|
protein phosphatase with EF hand calcium-binding domain 1 |
| chr19_-_38032006 | 0.14 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr15_-_55411560 | 0.14 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr4_-_118266416 | 0.14 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr4_+_11579648 | 0.13 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr4_+_118266582 | 0.13 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chrX_+_149377416 | 0.13 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_-_115236354 | 0.13 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr1_+_177272297 | 0.13 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr14_-_71003973 | 0.12 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr12_-_107969853 | 0.12 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr12_+_71063431 | 0.12 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_+_73142945 | 0.12 |
ENSMUST00000090811.11
ENSMUST00000112050.2 |
Scrn3
|
secernin 3 |
| chr9_-_105398346 | 0.11 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr1_+_87332638 | 0.11 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr17_+_46471950 | 0.11 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr8_-_45747883 | 0.10 |
ENSMUST00000026907.6
|
Klkb1
|
kallikrein B, plasma 1 |
| chr19_+_56414114 | 0.10 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr18_+_34891941 | 0.10 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr5_-_138169253 | 0.09 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr18_+_38809771 | 0.09 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr8_+_108271643 | 0.09 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_-_160079007 | 0.09 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr19_-_38031774 | 0.09 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr17_+_28226319 | 0.08 |
ENSMUST00000139173.3
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr18_+_34892599 | 0.08 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr6_-_136899167 | 0.07 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr11_+_54517164 | 0.07 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_87724512 | 0.07 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_+_47816137 | 0.07 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr5_+_104447037 | 0.07 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr5_+_88731386 | 0.07 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_+_4305273 | 0.07 |
ENSMUST00000175669.8
|
Frmd4a
|
FERM domain containing 4A |
| chr18_+_69652837 | 0.07 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr14_-_71004019 | 0.07 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr4_+_97665992 | 0.06 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr1_+_155911451 | 0.06 |
ENSMUST00000111754.9
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr12_-_107969673 | 0.06 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr9_+_47441471 | 0.06 |
ENSMUST00000114548.8
ENSMUST00000152459.8 ENSMUST00000143026.9 ENSMUST00000085909.9 ENSMUST00000114547.8 ENSMUST00000239368.2 ENSMUST00000214542.2 ENSMUST00000034581.4 |
Cadm1
|
cell adhesion molecule 1 |
| chr18_+_69652880 | 0.06 |
ENSMUST00000200813.4
|
Tcf4
|
transcription factor 4 |
| chr10_-_14593935 | 0.06 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr9_+_123635529 | 0.06 |
ENSMUST00000049810.9
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr13_+_94954202 | 0.06 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr18_+_69652550 | 0.06 |
ENSMUST00000201205.4
|
Tcf4
|
transcription factor 4 |
| chr8_+_112262729 | 0.06 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr2_-_118558825 | 0.06 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr2_-_70885877 | 0.06 |
ENSMUST00000090849.6
ENSMUST00000100037.9 ENSMUST00000112186.9 |
Mettl8
|
methyltransferase like 8 |
| chr4_-_82803384 | 0.05 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr3_+_135531409 | 0.05 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr10_+_59942274 | 0.05 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr11_-_107238956 | 0.05 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr16_+_17149235 | 0.05 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr9_+_123635556 | 0.05 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr17_-_80203457 | 0.05 |
ENSMUST00000068282.7
ENSMUST00000112437.8 |
Atl2
|
atlastin GTPase 2 |
| chr13_-_100753419 | 0.05 |
ENSMUST00000168772.2
ENSMUST00000163163.9 ENSMUST00000022137.14 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr12_-_32258469 | 0.05 |
ENSMUST00000085469.6
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr12_-_32258331 | 0.05 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr6_+_15185202 | 0.05 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr4_-_127864744 | 0.04 |
ENSMUST00000030614.3
|
CK137956
|
cDNA sequence CK137956 |
| chr1_+_155911879 | 0.04 |
ENSMUST00000128941.8
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr11_-_69553390 | 0.04 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr18_-_84104574 | 0.04 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr3_+_85946145 | 0.04 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr3_-_146487102 | 0.04 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr1_+_58841808 | 0.04 |
ENSMUST00000190213.2
|
Casp8
|
caspase 8 |
| chr10_+_41179966 | 0.04 |
ENSMUST00000173494.4
|
Ak9
|
adenylate kinase 9 |
| chr2_+_87853118 | 0.04 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr14_+_48358331 | 0.04 |
ENSMUST00000226513.2
|
Peli2
|
pellino 2 |
| chr8_-_18791557 | 0.04 |
ENSMUST00000033846.7
|
Angpt2
|
angiopoietin 2 |
| chr17_+_47816074 | 0.03 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr5_-_123127148 | 0.03 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr2_+_109522781 | 0.03 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr18_+_69652751 | 0.03 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr9_+_70114623 | 0.03 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chr14_-_50538979 | 0.03 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr5_-_18093739 | 0.03 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr18_-_84104507 | 0.03 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr3_+_68491487 | 0.03 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr17_+_38143840 | 0.03 |
ENSMUST00000213857.2
|
Olfr125
|
olfactory receptor 125 |
| chr9_-_101076198 | 0.03 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_-_99994257 | 0.03 |
ENSMUST00000207634.2
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr5_-_130284366 | 0.03 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr19_+_5091365 | 0.03 |
ENSMUST00000116567.4
ENSMUST00000235918.2 |
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr3_+_125197722 | 0.02 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr9_+_37656402 | 0.02 |
ENSMUST00000216982.2
|
Olfr874
|
olfactory receptor 874 |
| chr2_+_91560472 | 0.02 |
ENSMUST00000099712.10
ENSMUST00000111317.9 ENSMUST00000111316.9 ENSMUST00000045705.14 |
Ambra1
|
autophagy/beclin 1 regulator 1 |
| chr1_+_51328265 | 0.02 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr2_-_7400690 | 0.02 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_-_34882917 | 0.02 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr6_-_134768288 | 0.02 |
ENSMUST00000149776.3
|
Dusp16
|
dual specificity phosphatase 16 |
| chr18_-_75094323 | 0.02 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr3_-_9029097 | 0.02 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr2_+_91376650 | 0.02 |
ENSMUST00000099716.11
ENSMUST00000046769.16 ENSMUST00000111337.3 |
Ckap5
|
cytoskeleton associated protein 5 |
| chr7_+_44498640 | 0.02 |
ENSMUST00000054343.15
ENSMUST00000142880.3 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr3_+_85878376 | 0.02 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_-_118558852 | 0.02 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr3_-_20329823 | 0.02 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr1_-_184543367 | 0.02 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chr5_-_104261556 | 0.02 |
ENSMUST00000031249.8
|
Sparcl1
|
SPARC-like 1 |
| chr12_+_110452143 | 0.02 |
ENSMUST00000221715.2
ENSMUST00000109832.3 |
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_-_131918926 | 0.02 |
ENSMUST00000080215.6
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr8_-_64659004 | 0.01 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr17_-_71158703 | 0.01 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr1_-_97904958 | 0.01 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr18_+_37827413 | 0.01 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr1_-_75119277 | 0.01 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr11_-_99877423 | 0.01 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr19_-_11291805 | 0.01 |
ENSMUST00000187467.2
|
Ms4a14
|
membrane-spanning 4-domains, subfamily A, member 14 |
| chr12_-_81014849 | 0.01 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr13_+_38335232 | 0.01 |
ENSMUST00000124830.3
|
Dsp
|
desmoplakin |
| chr2_-_51863203 | 0.01 |
ENSMUST00000028314.9
|
Nmi
|
N-myc (and STAT) interactor |
| chr6_+_30541581 | 0.01 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr3_+_53396120 | 0.01 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chrX_+_162694397 | 0.01 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr15_-_5137951 | 0.01 |
ENSMUST00000141020.2
|
Card6
|
caspase recruitment domain family, member 6 |
| chr6_+_34686543 | 0.01 |
ENSMUST00000031775.13
|
Cald1
|
caldesmon 1 |
| chr6_+_34840057 | 0.01 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr4_-_58499398 | 0.01 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr6_+_17491231 | 0.01 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr17_+_47815968 | 0.01 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr3_-_154760978 | 0.01 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr12_+_38830812 | 0.01 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr3_-_102690026 | 0.01 |
ENSMUST00000172026.8
ENSMUST00000170856.6 |
Tshb
|
thyroid stimulating hormone, beta subunit |
| chr12_-_32000534 | 0.01 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr7_+_44498415 | 0.01 |
ENSMUST00000107885.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chrX_+_56257374 | 0.01 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr15_-_3333003 | 0.01 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr1_+_156666485 | 0.01 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr14_-_61495934 | 0.00 |
ENSMUST00000077954.13
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein) |
| chr7_+_113365235 | 0.00 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr4_+_34893772 | 0.00 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr10_+_87697155 | 0.00 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr11_+_99748741 | 0.00 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr12_+_113038376 | 0.00 |
ENSMUST00000109729.3
|
Tex22
|
testis expressed gene 22 |
| chr2_-_17465410 | 0.00 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr11_-_99695272 | 0.00 |
ENSMUST00000105056.2
|
Gm11554
|
predicted gene 11554 |
| chr11_-_16958647 | 0.00 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr11_-_99706484 | 0.00 |
ENSMUST00000105055.2
|
Gm11564
|
predicted gene 11564 |
| chr6_-_41535292 | 0.00 |
ENSMUST00000103300.3
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr3_+_75982890 | 0.00 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr9_+_43983493 | 0.00 |
ENSMUST00000176671.2
|
Usp2
|
ubiquitin specific peptidase 2 |
| chrX_+_162923474 | 0.00 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr12_-_81014755 | 0.00 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr6_-_41535322 | 0.00 |
ENSMUST00000193003.2
|
Trbv31
|
T cell receptor beta, variable 31 |
| chr5_-_90788323 | 0.00 |
ENSMUST00000202784.4
ENSMUST00000031317.10 ENSMUST00000201370.2 |
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_-_86944911 | 0.00 |
ENSMUST00000216088.3
|
Olfr259
|
olfactory receptor 259 |
| chrX_+_158086253 | 0.00 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr2_-_89882666 | 0.00 |
ENSMUST00000099757.2
|
Olfr140
|
olfactory receptor 140 |
| chr17_-_12894716 | 0.00 |
ENSMUST00000024596.10
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.4 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.0 | GO:0009216 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0070095 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |