Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
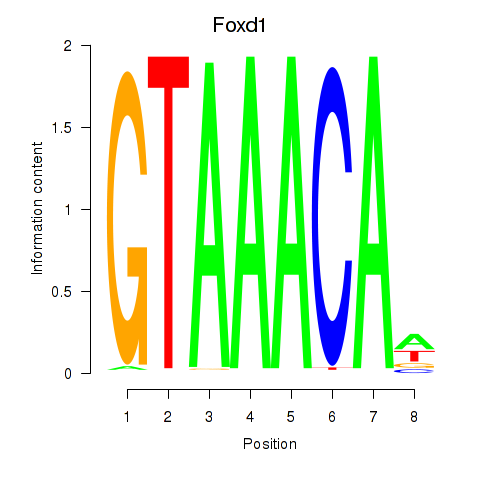
Results for Foxd1
Z-value: 0.20
Transcription factors associated with Foxd1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd1
|
ENSMUSG00000078302.6 | forkhead box D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd1 | mm39_v1_chr13_+_98490742_98490758 | -0.11 | 8.6e-01 | Click! |
Activity profile of Foxd1 motif
Sorted Z-values of Foxd1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102255999 | 0.16 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr11_+_68986043 | 0.12 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr6_-_124888643 | 0.12 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr5_-_138169476 | 0.12 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr14_+_48358267 | 0.10 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chrX_-_139443926 | 0.08 |
ENSMUST00000055738.12
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr7_-_25488060 | 0.08 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr15_-_79430742 | 0.08 |
ENSMUST00000231053.2
ENSMUST00000229431.2 |
Ddx17
|
DEAD box helicase 17 |
| chr14_+_54496683 | 0.07 |
ENSMUST00000041197.13
ENSMUST00000239403.2 ENSMUST00000197605.3 |
Abhd4
|
abhydrolase domain containing 4 |
| chrX_+_168468186 | 0.07 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr1_+_173501215 | 0.07 |
ENSMUST00000085876.12
|
Ifi208
|
interferon activated gene 208 |
| chr11_+_3280401 | 0.07 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr19_+_45433899 | 0.06 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr11_+_3282424 | 0.05 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_-_71198091 | 0.05 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr8_+_106785434 | 0.05 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr5_+_139375540 | 0.05 |
ENSMUST00000100514.3
|
Gpr146
|
G protein-coupled receptor 146 |
| chr11_+_3280771 | 0.05 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_102965910 | 0.05 |
ENSMUST00000199367.5
ENSMUST00000199049.5 ENSMUST00000197678.5 |
Nras
|
neuroblastoma ras oncogene |
| chr19_+_7534816 | 0.05 |
ENSMUST00000136465.8
ENSMUST00000025925.11 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr18_+_69477541 | 0.05 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr6_+_34575435 | 0.05 |
ENSMUST00000079391.10
ENSMUST00000142512.8 ENSMUST00000115027.8 ENSMUST00000115026.8 |
Cald1
|
caldesmon 1 |
| chr14_-_71003973 | 0.05 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr15_-_79430942 | 0.05 |
ENSMUST00000054014.9
ENSMUST00000229877.2 |
Ddx17
|
DEAD box helicase 17 |
| chr12_+_71063431 | 0.05 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr4_-_129534403 | 0.04 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr3_+_129326004 | 0.04 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr11_+_98632696 | 0.04 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr1_-_64995982 | 0.04 |
ENSMUST00000097713.2
|
Plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr18_+_34892599 | 0.04 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr9_+_119231140 | 0.04 |
ENSMUST00000165044.3
|
Acvr2b
|
activin receptor IIB |
| chr11_+_120123727 | 0.04 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr1_+_171052623 | 0.04 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr18_+_34891941 | 0.04 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr11_-_20691440 | 0.04 |
ENSMUST00000177543.8
|
Aftph
|
aftiphilin |
| chr3_-_27207931 | 0.04 |
ENSMUST00000175857.2
ENSMUST00000177055.8 ENSMUST00000176535.8 |
Ect2
|
ect2 oncogene |
| chr5_+_21577640 | 0.04 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr6_-_83808717 | 0.04 |
ENSMUST00000058383.9
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr19_+_46611826 | 0.04 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr14_-_71004019 | 0.04 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr10_+_59942274 | 0.04 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr6_+_134807097 | 0.04 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr17_+_85265420 | 0.04 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr17_+_29712008 | 0.04 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chrX_+_100298134 | 0.04 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr9_+_7272514 | 0.04 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr17_+_34251322 | 0.04 |
ENSMUST00000116612.3
|
Rxrb
|
retinoid X receptor beta |
| chr18_+_69652837 | 0.04 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr2_+_28531239 | 0.04 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr10_+_79951613 | 0.03 |
ENSMUST00000003152.14
|
Stk11
|
serine/threonine kinase 11 |
| chr18_+_36797113 | 0.03 |
ENSMUST00000036765.8
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr6_+_15185202 | 0.03 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr14_+_4230569 | 0.03 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr18_+_69652550 | 0.03 |
ENSMUST00000201205.4
|
Tcf4
|
transcription factor 4 |
| chr14_+_4230658 | 0.03 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr7_+_141056305 | 0.03 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr5_-_138169253 | 0.03 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr3_-_27207993 | 0.03 |
ENSMUST00000176242.9
ENSMUST00000176780.8 |
Ect2
|
ect2 oncogene |
| chr19_+_7534838 | 0.03 |
ENSMUST00000141887.8
ENSMUST00000136756.2 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr16_-_22258469 | 0.03 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr15_-_98816012 | 0.03 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr12_+_35097179 | 0.03 |
ENSMUST00000163677.3
ENSMUST00000048519.17 |
Snx13
|
sorting nexin 13 |
| chr17_+_34251041 | 0.03 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta |
| chr7_-_143239600 | 0.03 |
ENSMUST00000208017.2
ENSMUST00000152703.2 |
Tnfrsf23
|
tumor necrosis factor receptor superfamily, member 23 |
| chr1_+_173093568 | 0.03 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr8_-_85500010 | 0.03 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr7_+_24607042 | 0.03 |
ENSMUST00000151121.2
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr1_+_34044940 | 0.03 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr1_+_52158721 | 0.03 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr19_-_29721012 | 0.03 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr13_-_93636224 | 0.03 |
ENSMUST00000220513.2
ENSMUST00000065537.9 |
Jmy
|
junction-mediating and regulatory protein |
| chr2_+_73142945 | 0.03 |
ENSMUST00000090811.11
ENSMUST00000112050.2 |
Scrn3
|
secernin 3 |
| chr4_-_149760488 | 0.03 |
ENSMUST00000118704.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr6_-_87786736 | 0.03 |
ENSMUST00000032134.9
|
Rab43
|
RAB43, member RAS oncogene family |
| chr5_+_107585774 | 0.03 |
ENSMUST00000162298.4
ENSMUST00000094541.4 ENSMUST00000211896.2 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr5_-_142803405 | 0.03 |
ENSMUST00000151477.8
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr4_+_144619696 | 0.03 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr1_+_52158693 | 0.03 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_+_52158599 | 0.03 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr2_+_129642371 | 0.02 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr7_+_16044336 | 0.02 |
ENSMUST00000136781.2
|
Bbc3
|
BCL2 binding component 3 |
| chr8_-_3329605 | 0.02 |
ENSMUST00000091291.5
|
Insr
|
insulin receptor |
| chr10_-_93146825 | 0.02 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr9_+_96078340 | 0.02 |
ENSMUST00000034982.16
ENSMUST00000188008.7 ENSMUST00000188750.7 ENSMUST00000185644.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr19_-_7319211 | 0.02 |
ENSMUST00000171393.8
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr16_-_22258320 | 0.02 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr5_-_138169509 | 0.02 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr1_-_138547879 | 0.02 |
ENSMUST00000187407.7
|
Nek7
|
NIMA (never in mitosis gene a)-related expressed kinase 7 |
| chr1_-_138547403 | 0.02 |
ENSMUST00000027642.5
ENSMUST00000186017.7 |
Nek7
|
NIMA (never in mitosis gene a)-related expressed kinase 7 |
| chr15_+_6451721 | 0.02 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr19_+_56414114 | 0.02 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr3_-_37286714 | 0.02 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr5_+_31409021 | 0.02 |
ENSMUST00000054829.13
ENSMUST00000201625.4 ENSMUST00000201937.4 |
Krtcap3
|
keratinocyte associated protein 3 |
| chr13_+_104315301 | 0.02 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chr17_+_47816137 | 0.02 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr11_-_76386190 | 0.02 |
ENSMUST00000108408.9
|
Abr
|
active BCR-related gene |
| chr6_+_29853745 | 0.02 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr18_+_69652880 | 0.02 |
ENSMUST00000200813.4
|
Tcf4
|
transcription factor 4 |
| chr11_-_23448030 | 0.02 |
ENSMUST00000020529.13
|
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr17_-_48189815 | 0.02 |
ENSMUST00000154108.2
|
Foxp4
|
forkhead box P4 |
| chr15_+_8997480 | 0.02 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr10_+_29087602 | 0.02 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr19_-_27988393 | 0.02 |
ENSMUST00000172907.8
ENSMUST00000046898.17 |
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr1_-_183078488 | 0.02 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr14_-_68893253 | 0.02 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr14_+_48358331 | 0.02 |
ENSMUST00000226513.2
|
Peli2
|
pellino 2 |
| chr18_-_39622829 | 0.02 |
ENSMUST00000025300.13
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr3_-_146475974 | 0.02 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr2_+_24866039 | 0.02 |
ENSMUST00000045295.14
|
Pnpla7
|
patatin-like phospholipase domain containing 7 |
| chr3_-_61273228 | 0.02 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr4_+_11191354 | 0.02 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr9_+_102593871 | 0.02 |
ENSMUST00000145913.2
ENSMUST00000153965.8 |
Amotl2
|
angiomotin-like 2 |
| chr11_-_107238956 | 0.02 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr18_+_77153332 | 0.02 |
ENSMUST00000114776.4
ENSMUST00000168882.9 |
Pias2
|
protein inhibitor of activated STAT 2 |
| chr15_-_80951210 | 0.02 |
ENSMUST00000239267.2
|
Mrtfa
|
myocardin related transcription factor A |
| chr17_+_25105617 | 0.02 |
ENSMUST00000117890.8
ENSMUST00000168265.8 ENSMUST00000120943.8 ENSMUST00000068508.13 ENSMUST00000119829.8 |
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr14_+_55798362 | 0.01 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr19_-_45771939 | 0.01 |
ENSMUST00000026243.5
|
Oga
|
O-GlcNAcase |
| chr6_-_71417607 | 0.01 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr13_-_53083494 | 0.01 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr1_+_179373935 | 0.01 |
ENSMUST00000040706.9
|
Cnst
|
consortin, connexin sorting protein |
| chr13_+_55357585 | 0.01 |
ENSMUST00000224973.2
ENSMUST00000099490.3 |
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr11_+_70104929 | 0.01 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr3_+_138566249 | 0.01 |
ENSMUST00000121826.3
|
Tspan5
|
tetraspanin 5 |
| chr13_+_118851214 | 0.01 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr19_+_13890894 | 0.01 |
ENSMUST00000216623.2
ENSMUST00000216835.2 |
Olfr1505
|
olfactory receptor 1505 |
| chr12_-_83643883 | 0.01 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr3_-_27208010 | 0.01 |
ENSMUST00000108300.8
ENSMUST00000108298.9 |
Ect2
|
ect2 oncogene |
| chr3_+_102965601 | 0.01 |
ENSMUST00000029445.13
ENSMUST00000200069.5 |
Nras
|
neuroblastoma ras oncogene |
| chr17_+_34250757 | 0.01 |
ENSMUST00000044858.16
ENSMUST00000174299.9 |
Rxrb
|
retinoid X receptor beta |
| chr4_+_11191726 | 0.01 |
ENSMUST00000029866.16
ENSMUST00000108324.4 |
Ccne2
|
cyclin E2 |
| chr19_+_5927821 | 0.01 |
ENSMUST00000145200.8
ENSMUST00000025732.14 ENSMUST00000125114.8 ENSMUST00000155697.8 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr15_+_102012782 | 0.01 |
ENSMUST00000230474.2
|
Tns2
|
tensin 2 |
| chr13_-_96803216 | 0.01 |
ENSMUST00000170287.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr8_+_35274440 | 0.01 |
ENSMUST00000033930.5
|
Dusp4
|
dual specificity phosphatase 4 |
| chr4_-_45012287 | 0.01 |
ENSMUST00000055028.9
ENSMUST00000180217.2 ENSMUST00000107817.3 |
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr4_-_45012093 | 0.01 |
ENSMUST00000131991.2
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr13_+_55593116 | 0.01 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr3_+_96465265 | 0.01 |
ENSMUST00000074519.13
ENSMUST00000049093.8 |
Txnip
|
thioredoxin interacting protein |
| chr19_+_34268071 | 0.01 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr4_+_132857816 | 0.01 |
ENSMUST00000084241.12
ENSMUST00000138831.2 |
Wasf2
|
WASP family, member 2 |
| chr6_-_148847854 | 0.01 |
ENSMUST00000139355.8
ENSMUST00000146457.2 ENSMUST00000054080.15 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr18_+_69652751 | 0.01 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr1_-_45542442 | 0.01 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr6_+_121323577 | 0.01 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr12_-_85317359 | 0.01 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr14_+_55798517 | 0.01 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_-_180023518 | 0.01 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr3_+_129326285 | 0.01 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr9_-_45866264 | 0.01 |
ENSMUST00000114573.9
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr14_-_54651442 | 0.01 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr2_-_103858632 | 0.01 |
ENSMUST00000056170.4
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr4_+_134042423 | 0.01 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr11_-_101117237 | 0.01 |
ENSMUST00000100417.3
ENSMUST00000107285.8 ENSMUST00000107284.8 |
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr4_+_155819257 | 0.01 |
ENSMUST00000147721.8
ENSMUST00000127188.3 |
Tmem240
|
transmembrane protein 240 |
| chr15_+_59520199 | 0.01 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr6_-_148847633 | 0.01 |
ENSMUST00000132696.8
|
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr2_+_181479647 | 0.01 |
ENSMUST00000029116.14
ENSMUST00000108754.8 |
Pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr2_+_38401826 | 0.01 |
ENSMUST00000112895.8
|
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr14_-_40730180 | 0.01 |
ENSMUST00000136661.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr9_+_96078299 | 0.01 |
ENSMUST00000165120.9
|
Tfdp2
|
transcription factor Dp 2 |
| chr16_+_24266829 | 0.01 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr3_+_53396120 | 0.01 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr1_-_80191649 | 0.01 |
ENSMUST00000058748.2
|
Fam124b
|
family with sequence similarity 124, member B |
| chr9_-_101076198 | 0.01 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr10_+_115653152 | 0.01 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr2_+_121859025 | 0.01 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr11_-_23447866 | 0.01 |
ENSMUST00000128559.2
ENSMUST00000147157.8 ENSMUST00000109539.8 |
Ahsa2
|
AHA1, activator of heat shock protein ATPase 2 |
| chr19_-_27988534 | 0.01 |
ENSMUST00000174850.8
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr11_-_107361525 | 0.01 |
ENSMUST00000103064.10
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_+_42734889 | 0.01 |
ENSMUST00000054883.4
|
Pou3f3
|
POU domain, class 3, transcription factor 3 |
| chr14_-_31503869 | 0.01 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr3_+_32490300 | 0.01 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr12_-_32000534 | 0.01 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr3_+_32490525 | 0.01 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr12_+_84498196 | 0.01 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr1_+_36550948 | 0.01 |
ENSMUST00000001166.14
ENSMUST00000097776.4 |
Cnnm3
|
cyclin M3 |
| chr18_-_39622295 | 0.01 |
ENSMUST00000131885.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_-_53083674 | 0.01 |
ENSMUST00000120535.8
ENSMUST00000119311.8 ENSMUST00000021913.16 ENSMUST00000110031.4 |
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr3_+_90161470 | 0.01 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr9_+_102594867 | 0.01 |
ENSMUST00000035121.14
|
Amotl2
|
angiomotin-like 2 |
| chr6_-_52135261 | 0.01 |
ENSMUST00000000964.6
ENSMUST00000120363.2 |
Hoxa1
|
homeobox A1 |
| chr8_-_84963653 | 0.01 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr5_-_137530214 | 0.01 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_+_183078573 | 0.01 |
ENSMUST00000109166.8
|
Aida
|
axin interactor, dorsalization associated |
| chr10_-_80513927 | 0.01 |
ENSMUST00000199949.2
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr11_-_120881697 | 0.01 |
ENSMUST00000154483.8
|
Csnk1d
|
casein kinase 1, delta |
| chr17_+_47816074 | 0.01 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr8_+_47192911 | 0.01 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chr13_-_63712349 | 0.01 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr8_+_47193275 | 0.01 |
ENSMUST00000207571.3
|
Irf2
|
interferon regulatory factor 2 |
| chr6_-_106777014 | 0.01 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr11_+_110914678 | 0.01 |
ENSMUST00000150902.8
ENSMUST00000178798.2 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_-_41836514 | 0.01 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr5_-_137529465 | 0.01 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_-_64995891 | 0.01 |
ENSMUST00000123225.2
|
Plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr4_-_117146624 | 0.01 |
ENSMUST00000221654.2
|
Rnf220
|
ring finger protein 220 |
| chr11_-_119977609 | 0.01 |
ENSMUST00000106227.8
ENSMUST00000106229.8 ENSMUST00000180242.2 |
Cep131
|
centrosomal protein 131 |
| chr10_-_85020867 | 0.01 |
ENSMUST00000020227.11
|
Cry1
|
cryptochrome 1 (photolyase-like) |
| chr9_-_108526548 | 0.01 |
ENSMUST00000013338.14
ENSMUST00000193197.6 |
Arih2
|
ariadne RBR E3 ubiquitin protein ligase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxd1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0097048 | dendritic cell apoptotic process(GO:0097048) neutrophil clearance(GO:0097350) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.0 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |