Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
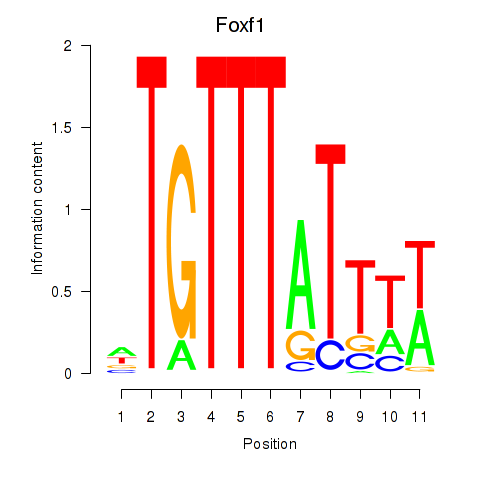
Results for Foxf1
Z-value: 0.60
Transcription factors associated with Foxf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf1
|
ENSMUSG00000042812.6 | forkhead box F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf1 | mm39_v1_chr8_+_121811091_121811125 | -0.27 | 6.6e-01 | Click! |
Activity profile of Foxf1 motif
Sorted Z-values of Foxf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_61844899 | 0.37 |
ENSMUST00000225582.2
ENSMUST00000051184.10 |
Kcnrg
|
potassium channel regulator |
| chr5_+_146321757 | 0.30 |
ENSMUST00000016143.9
|
Wasf3
|
WASP family, member 3 |
| chr7_-_103463120 | 0.30 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr5_+_88731386 | 0.30 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_-_87868043 | 0.29 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr9_-_39486831 | 0.25 |
ENSMUST00000216298.2
ENSMUST00000215194.2 |
Olfr959
|
olfactory receptor 959 |
| chr13_-_32967937 | 0.25 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr5_+_104447037 | 0.24 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr10_-_12745109 | 0.23 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr11_-_54751738 | 0.22 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr7_+_16807965 | 0.22 |
ENSMUST00000071399.13
ENSMUST00000118367.3 |
Psg16
|
pregnancy specific glycoprotein 16 |
| chr11_-_110058899 | 0.21 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr8_-_64659004 | 0.20 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr18_-_60724855 | 0.20 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr16_+_38562616 | 0.20 |
ENSMUST00000023482.13
ENSMUST00000114712.8 ENSMUST00000231655.2 |
B4galt4
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 4 |
| chr2_-_144369261 | 0.19 |
ENSMUST00000163701.2
ENSMUST00000081982.12 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr11_+_101473062 | 0.19 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr15_-_5137975 | 0.19 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr2_+_4305273 | 0.18 |
ENSMUST00000175669.8
|
Frmd4a
|
FERM domain containing 4A |
| chr2_+_113235475 | 0.18 |
ENSMUST00000238883.2
|
Fmn1
|
formin 1 |
| chr6_+_43149074 | 0.17 |
ENSMUST00000216179.2
|
Olfr13
|
olfactory receptor 13 |
| chr16_-_43484494 | 0.17 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr1_+_37338964 | 0.17 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr18_+_37580692 | 0.16 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr4_-_82803384 | 0.16 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr3_-_10273628 | 0.16 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr10_-_14593935 | 0.16 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr11_-_65636651 | 0.15 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr8_-_65582206 | 0.15 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr17_+_29077385 | 0.15 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr10_+_129153986 | 0.14 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780 |
| chr11_+_97732108 | 0.14 |
ENSMUST00000155954.3
ENSMUST00000164364.2 ENSMUST00000170806.2 |
B230217C12Rik
|
RIKEN cDNA B230217C12 gene |
| chr15_+_6673167 | 0.14 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr3_+_53396120 | 0.14 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr13_-_96678987 | 0.14 |
ENSMUST00000022172.12
|
Polk
|
polymerase (DNA directed), kappa |
| chr13_+_23317325 | 0.13 |
ENSMUST00000227050.2
ENSMUST00000227160.2 ENSMUST00000227741.2 ENSMUST00000226692.2 |
Vmn1r218
|
vomeronasal 1 receptor 218 |
| chr13_+_23191826 | 0.13 |
ENSMUST00000228758.2
ENSMUST00000228031.2 ENSMUST00000227573.2 |
Vmn1r213
|
vomeronasal 1 receptor 213 |
| chr14_-_50538979 | 0.13 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr7_-_5548308 | 0.13 |
ENSMUST00000236262.2
|
Vmn1r60
|
vomeronasal 1 receptor 60 |
| chr1_+_63655127 | 0.12 |
ENSMUST00000226288.2
|
Gm39653
|
predicted gene, 39653 |
| chr3_-_64473251 | 0.12 |
ENSMUST00000176481.9
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
| chr3_+_33853941 | 0.12 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr1_-_87322443 | 0.12 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr19_+_38252984 | 0.12 |
ENSMUST00000198518.5
ENSMUST00000199812.5 |
Lgi1
|
leucine-rich repeat LGI family, member 1 |
| chr16_-_85347305 | 0.12 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr2_-_90309509 | 0.12 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr13_-_30039613 | 0.12 |
ENSMUST00000091674.12
ENSMUST00000006353.14 |
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr2_+_71884943 | 0.11 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_50586329 | 0.11 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr16_+_10363254 | 0.11 |
ENSMUST00000115827.8
ENSMUST00000150894.2 ENSMUST00000038145.13 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr6_+_146934082 | 0.11 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr8_-_87611849 | 0.11 |
ENSMUST00000034074.8
|
N4bp1
|
NEDD4 binding protein 1 |
| chr12_-_11315755 | 0.11 |
ENSMUST00000166117.4
ENSMUST00000219600.2 ENSMUST00000218487.2 |
Gen1
|
GEN1, Holliday junction 5' flap endonuclease |
| chr9_+_65583852 | 0.10 |
ENSMUST00000136166.3
|
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr6_+_68098030 | 0.10 |
ENSMUST00000103317.3
|
Igkv1-117
|
immunoglobulin kappa variable 1-117 |
| chr14_+_54664359 | 0.10 |
ENSMUST00000010550.12
ENSMUST00000199195.3 ENSMUST00000196273.2 |
Mrpl52
|
mitochondrial ribosomal protein L52 |
| chr10_+_129219952 | 0.10 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr1_+_171052623 | 0.10 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr2_-_51039112 | 0.10 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr18_-_80751327 | 0.10 |
ENSMUST00000236310.2
ENSMUST00000167977.8 ENSMUST00000035800.8 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr18_-_46345661 | 0.10 |
ENSMUST00000037011.6
|
Trim36
|
tripartite motif-containing 36 |
| chr19_+_13595285 | 0.09 |
ENSMUST00000216688.3
|
Olfr1487
|
olfactory receptor 1487 |
| chr19_-_17814984 | 0.09 |
ENSMUST00000025618.16
ENSMUST00000050715.10 |
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr2_+_91560472 | 0.09 |
ENSMUST00000099712.10
ENSMUST00000111317.9 ENSMUST00000111316.9 ENSMUST00000045705.14 |
Ambra1
|
autophagy/beclin 1 regulator 1 |
| chr6_+_129510331 | 0.09 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr17_+_37670473 | 0.09 |
ENSMUST00000178766.3
ENSMUST00000215398.2 |
Olfr104-ps
|
olfactory receptor 104, pseudogene |
| chr13_+_23398297 | 0.09 |
ENSMUST00000236177.2
|
Vmn1r221
|
vomeronasal 1 receptor 221 |
| chr2_-_111908072 | 0.09 |
ENSMUST00000213577.2
ENSMUST00000216071.2 |
Olfr1313
|
olfactory receptor 1313 |
| chr2_-_86257093 | 0.09 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062 |
| chr2_-_86208737 | 0.09 |
ENSMUST00000217435.2
|
Olfr1057
|
olfactory receptor 1057 |
| chr19_+_43770619 | 0.08 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr6_-_113696809 | 0.08 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr12_+_71095112 | 0.08 |
ENSMUST00000135709.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr13_+_83672654 | 0.08 |
ENSMUST00000199019.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_160079007 | 0.08 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr5_-_98178834 | 0.08 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2 |
| chr14_-_50479161 | 0.07 |
ENSMUST00000214388.2
|
Olfr731
|
olfactory receptor 731 |
| chr16_-_55755208 | 0.07 |
ENSMUST00000121129.8
ENSMUST00000023270.14 |
Cep97
|
centrosomal protein 97 |
| chr6_-_136852792 | 0.07 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr2_-_87838612 | 0.07 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr13_+_83672708 | 0.07 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr16_-_55755160 | 0.07 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
| chr1_-_169938060 | 0.07 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr17_+_17669082 | 0.07 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr7_-_101714251 | 0.07 |
ENSMUST00000130074.2
ENSMUST00000131104.3 ENSMUST00000096639.12 |
Rnf121
|
ring finger protein 121 |
| chr17_+_33410276 | 0.07 |
ENSMUST00000214406.2
ENSMUST00000213731.2 |
Olfr239
|
olfactory receptor 239 |
| chr13_+_22656093 | 0.07 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr7_-_103674780 | 0.07 |
ENSMUST00000218535.2
|
Olfr640
|
olfactory receptor 640 |
| chr2_-_88581690 | 0.07 |
ENSMUST00000215179.3
ENSMUST00000215529.3 |
Olfr1198
|
olfactory receptor 1198 |
| chrX_+_7445806 | 0.07 |
ENSMUST00000234363.2
ENSMUST00000235116.2 ENSMUST00000115739.9 ENSMUST00000234574.2 ENSMUST00000115740.9 |
Foxp3
|
forkhead box P3 |
| chr14_-_50479323 | 0.07 |
ENSMUST00000214152.2
|
Olfr731
|
olfactory receptor 731 |
| chr19_-_6002210 | 0.07 |
ENSMUST00000236013.2
|
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr19_+_13385213 | 0.06 |
ENSMUST00000216910.3
|
Olfr1469
|
olfactory receptor 1469 |
| chr1_+_177272215 | 0.06 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_+_85715984 | 0.06 |
ENSMUST00000213441.3
|
Olfr1023
|
olfactory receptor 1023 |
| chr12_-_79343040 | 0.06 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr11_-_86884507 | 0.06 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr16_+_80997580 | 0.06 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr8_+_84262409 | 0.06 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr6_-_39702127 | 0.06 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr2_+_111355548 | 0.06 |
ENSMUST00000217845.3
|
Olfr1293-ps
|
olfactory receptor 1293, pseudogene |
| chr13_+_49697919 | 0.06 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr2_+_87853118 | 0.06 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr10_+_129072073 | 0.06 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr2_-_45000389 | 0.06 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_129428869 | 0.05 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr18_-_39051695 | 0.05 |
ENSMUST00000040647.11
|
Fgf1
|
fibroblast growth factor 1 |
| chr17_+_43671314 | 0.05 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr15_-_80989200 | 0.05 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr19_-_19088543 | 0.05 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chr1_-_139487951 | 0.05 |
ENSMUST00000023965.8
|
Cfhr1
|
complement factor H-related 1 |
| chr7_+_44866635 | 0.05 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr2_-_71198091 | 0.05 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr5_-_18093739 | 0.05 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr9_-_101076198 | 0.05 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr14_-_54646917 | 0.05 |
ENSMUST00000000984.9
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr7_+_110371811 | 0.05 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr9_+_111011327 | 0.05 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_+_123635529 | 0.05 |
ENSMUST00000049810.9
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr17_-_56312555 | 0.05 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr13_-_23650045 | 0.05 |
ENSMUST00000041674.14
ENSMUST00000110434.2 |
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr4_+_102427247 | 0.05 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr3_-_20329823 | 0.04 |
ENSMUST00000011607.6
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr9_+_123635556 | 0.04 |
ENSMUST00000216072.2
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr12_+_52746158 | 0.04 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr18_+_4994600 | 0.04 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
| chr9_+_122717536 | 0.04 |
ENSMUST00000063980.8
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr18_+_69652837 | 0.04 |
ENSMUST00000201410.4
ENSMUST00000202937.4 |
Tcf4
|
transcription factor 4 |
| chr2_-_34716199 | 0.04 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr5_-_21150470 | 0.04 |
ENSMUST00000197089.2
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr12_-_32258604 | 0.04 |
ENSMUST00000053215.14
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr1_-_160134873 | 0.04 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr13_+_36301331 | 0.04 |
ENSMUST00000021857.13
|
Fars2
|
phenylalanine-tRNA synthetase 2 (mitochondrial) |
| chr7_-_65020655 | 0.04 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr6_-_113696390 | 0.04 |
ENSMUST00000203588.2
ENSMUST00000204163.3 ENSMUST00000203363.3 |
Ghrl
|
ghrelin |
| chr13_+_83672389 | 0.04 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr5_+_107656810 | 0.04 |
ENSMUST00000160160.6
|
Gm42669
|
predicted gene 42669 |
| chr5_-_106606032 | 0.04 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr6_-_124410452 | 0.04 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr1_+_87332638 | 0.04 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr14_-_54648057 | 0.04 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr17_+_19724994 | 0.04 |
ENSMUST00000166081.3
ENSMUST00000231465.2 |
Vmn2r100
|
vomeronasal 2, receptor 100 |
| chr9_+_78020554 | 0.03 |
ENSMUST00000009972.6
ENSMUST00000117330.8 ENSMUST00000044551.8 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr1_-_132318039 | 0.03 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr7_+_108209994 | 0.03 |
ENSMUST00000209296.3
|
Olfr506
|
olfactory receptor 506 |
| chr1_+_75119472 | 0.03 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chrX_-_107877909 | 0.03 |
ENSMUST00000101283.4
ENSMUST00000150434.8 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr7_+_101714943 | 0.03 |
ENSMUST00000094130.4
ENSMUST00000084843.10 |
Xndc1
Xntrpc
|
Xrcc1 N-terminal domain containing 1 Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
| chr10_+_25308466 | 0.03 |
ENSMUST00000219224.2
ENSMUST00000219166.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr16_+_43184191 | 0.03 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_+_51328265 | 0.03 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr3_-_19319155 | 0.03 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr5_-_66211842 | 0.03 |
ENSMUST00000200852.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr10_+_94350687 | 0.03 |
ENSMUST00000065060.12
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr9_+_107457316 | 0.03 |
ENSMUST00000093785.6
|
Naa80
|
N(alpha)-acetyltransferase 80, NatH catalytic subunit |
| chr10_+_97315465 | 0.03 |
ENSMUST00000105287.11
|
Dcn
|
decorin |
| chr7_-_65020955 | 0.03 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr12_-_73160181 | 0.03 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr2_+_112114906 | 0.03 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr16_-_22475960 | 0.03 |
ENSMUST00000023578.14
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr12_-_76842263 | 0.03 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr15_+_102315579 | 0.03 |
ENSMUST00000169619.2
|
Sp1
|
trans-acting transcription factor 1 |
| chr14_+_26616514 | 0.03 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr18_+_69652751 | 0.03 |
ENSMUST00000200966.4
|
Tcf4
|
transcription factor 4 |
| chr10_+_127256736 | 0.03 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr11_-_114907019 | 0.03 |
ENSMUST00000106578.8
ENSMUST00000092463.7 |
Cd300ld2
|
CD300 molecule like family member D2 |
| chr12_+_79075924 | 0.03 |
ENSMUST00000039928.7
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr14_-_24537067 | 0.03 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr5_+_88731366 | 0.03 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_+_153300874 | 0.03 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr6_-_119925387 | 0.03 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr6_-_29164981 | 0.03 |
ENSMUST00000007993.16
|
Rbm28
|
RNA binding motif protein 28 |
| chr12_-_32258469 | 0.02 |
ENSMUST00000085469.6
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr6_+_129510145 | 0.02 |
ENSMUST00000204487.3
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr2_-_118558852 | 0.02 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr9_+_123603605 | 0.02 |
ENSMUST00000180093.2
|
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr10_-_53225909 | 0.02 |
ENSMUST00000220376.2
|
Cep85l
|
centrosomal protein 85-like |
| chr1_-_64160557 | 0.02 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr18_+_7905440 | 0.02 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil |
| chr12_-_108241597 | 0.02 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr6_+_29361408 | 0.02 |
ENSMUST00000156163.2
|
Calu
|
calumenin |
| chr17_+_38110779 | 0.02 |
ENSMUST00000215168.2
ENSMUST00000216478.2 |
Olfr124
|
olfactory receptor 124 |
| chr14_-_96756503 | 0.02 |
ENSMUST00000022666.9
|
Klhl1
|
kelch-like 1 |
| chr7_+_113365235 | 0.02 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr2_-_118558825 | 0.02 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr3_-_123179574 | 0.02 |
ENSMUST00000090371.14
ENSMUST00000174323.6 ENSMUST00000029759.16 |
Mettl14
|
methyltransferase like 14 |
| chr9_+_37656402 | 0.02 |
ENSMUST00000216982.2
|
Olfr874
|
olfactory receptor 874 |
| chr11_+_4936824 | 0.02 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr2_-_86016027 | 0.02 |
ENSMUST00000215138.3
|
Olfr52
|
olfactory receptor 52 |
| chr13_-_36301466 | 0.02 |
ENSMUST00000053265.8
|
Lyrm4
|
LYR motif containing 4 |
| chr7_+_79048884 | 0.02 |
ENSMUST00000137667.3
|
Fanci
|
Fanconi anemia, complementation group I |
| chr1_+_10127147 | 0.02 |
ENSMUST00000149214.3
|
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr12_-_32258331 | 0.02 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr7_-_100306160 | 0.02 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr9_-_90152759 | 0.02 |
ENSMUST00000041767.14
ENSMUST00000128874.3 |
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr5_-_115236354 | 0.02 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr1_+_156666485 | 0.01 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr14_+_71011744 | 0.01 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chrX_+_82898974 | 0.01 |
ENSMUST00000239269.2
|
Dmd
|
dystrophin, muscular dystrophy |
| chr15_+_4756657 | 0.01 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.0 | 0.2 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.0 | 0.1 | GO:2000506 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0046340 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0002654 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |