Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
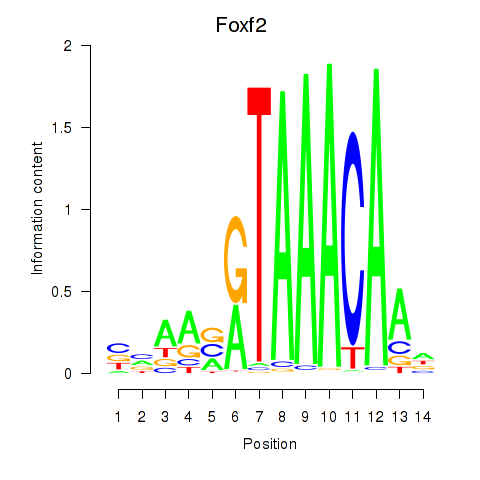
Results for Foxf2
Z-value: 0.46
Transcription factors associated with Foxf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxf2
|
ENSMUSG00000038402.3 | forkhead box F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf2 | mm39_v1_chr13_+_31809774_31809799 | -0.88 | 4.8e-02 | Click! |
Activity profile of Foxf2 motif
Sorted Z-values of Foxf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_106785434 | 0.26 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr4_-_87724533 | 0.23 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr17_+_29309942 | 0.21 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr5_+_65288418 | 0.21 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr17_+_47816137 | 0.20 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr19_-_4062738 | 0.17 |
ENSMUST00000136921.2
ENSMUST00000042497.14 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr3_-_143908111 | 0.17 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr3_-_143908060 | 0.17 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr4_-_87724512 | 0.16 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr2_+_164328375 | 0.16 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_-_4062656 | 0.16 |
ENSMUST00000134479.8
ENSMUST00000128787.8 ENSMUST00000237862.2 ENSMUST00000236203.2 ENSMUST00000133474.8 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr7_+_141056305 | 0.15 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr3_-_144412394 | 0.15 |
ENSMUST00000200532.2
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr5_+_86219593 | 0.15 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr17_+_47816042 | 0.15 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr17_+_47815968 | 0.14 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr17_+_47816074 | 0.14 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr3_+_65435825 | 0.14 |
ENSMUST00000047906.10
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr4_-_108690741 | 0.12 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr8_-_46577183 | 0.11 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr19_+_53317844 | 0.11 |
ENSMUST00000111737.3
ENSMUST00000025998.15 ENSMUST00000237837.2 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr10_+_87926932 | 0.10 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr15_-_98816012 | 0.10 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr5_-_115236354 | 0.10 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr5_-_92496730 | 0.09 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr12_+_69343450 | 0.09 |
ENSMUST00000021362.5
|
Klhdc2
|
kelch domain containing 2 |
| chr13_+_55593116 | 0.09 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr1_-_194813843 | 0.09 |
ENSMUST00000075451.12
ENSMUST00000191775.2 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr6_-_71417607 | 0.09 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr12_+_3857001 | 0.08 |
ENSMUST00000020991.15
ENSMUST00000172509.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chr7_+_81412695 | 0.08 |
ENSMUST00000133034.2
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr3_-_137837117 | 0.08 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr2_-_170248421 | 0.08 |
ENSMUST00000154650.8
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr13_+_44994167 | 0.07 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr12_-_25147139 | 0.07 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr7_+_81412673 | 0.07 |
ENSMUST00000042166.11
|
Ramac
|
RNA guanine-7 methyltransferase activating subunit |
| chr19_-_45772230 | 0.07 |
ENSMUST00000235448.2
|
Oga
|
O-GlcNAcase |
| chr1_-_97904958 | 0.07 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_-_66432938 | 0.07 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71 |
| chr19_+_45433899 | 0.07 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr3_+_85878376 | 0.07 |
ENSMUST00000238443.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_+_143757193 | 0.06 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr12_-_73160181 | 0.06 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr16_-_22258469 | 0.06 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr4_+_118266582 | 0.06 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr19_-_45771939 | 0.06 |
ENSMUST00000026243.5
|
Oga
|
O-GlcNAcase |
| chr2_-_25498459 | 0.06 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr1_+_165596961 | 0.06 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr18_-_88945571 | 0.06 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr3_+_65435878 | 0.05 |
ENSMUST00000130705.2
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr12_-_15866763 | 0.05 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr12_+_3857077 | 0.05 |
ENSMUST00000174817.8
|
Dnmt3a
|
DNA methyltransferase 3A |
| chr13_-_104056803 | 0.05 |
ENSMUST00000091269.11
ENSMUST00000188997.7 ENSMUST00000169083.8 ENSMUST00000191275.7 |
Erbin
|
Erbb2 interacting protein |
| chr2_+_164328763 | 0.05 |
ENSMUST00000109349.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_+_118266526 | 0.05 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr8_-_87472562 | 0.05 |
ENSMUST00000045296.6
|
Siah1a
|
siah E3 ubiquitin protein ligase 1A |
| chr9_-_48747232 | 0.05 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr1_-_179373826 | 0.05 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr1_-_180023518 | 0.04 |
ENSMUST00000162769.8
ENSMUST00000161379.2 ENSMUST00000027766.13 ENSMUST00000161814.8 |
Coq8a
|
coenzyme Q8A |
| chr1_+_75119419 | 0.04 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr12_+_98594388 | 0.04 |
ENSMUST00000048402.12
ENSMUST00000101144.10 ENSMUST00000101146.4 |
Spata7
|
spermatogenesis associated 7 |
| chr2_-_71198091 | 0.04 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr5_+_88731366 | 0.04 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr12_+_112645237 | 0.04 |
ENSMUST00000174780.2
ENSMUST00000169593.2 ENSMUST00000173942.2 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr10_-_37014859 | 0.04 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr2_-_118558825 | 0.04 |
ENSMUST00000159756.2
|
Plcb2
|
phospholipase C, beta 2 |
| chr9_+_110162470 | 0.04 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr8_+_112262729 | 0.03 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr2_-_86109346 | 0.03 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr6_+_34840057 | 0.03 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr14_+_27598021 | 0.03 |
ENSMUST00000211684.2
ENSMUST00000210924.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr6_+_146934082 | 0.03 |
ENSMUST00000036194.6
|
Rep15
|
RAB15 effector protein |
| chr4_+_5724305 | 0.03 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr7_-_141925947 | 0.03 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr14_+_65612788 | 0.03 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr19_-_11301919 | 0.03 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr8_+_70826191 | 0.02 |
ENSMUST00000003659.9
|
Comp
|
cartilage oligomeric matrix protein |
| chr19_-_38113696 | 0.02 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr4_+_101365144 | 0.02 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr6_-_13677928 | 0.02 |
ENSMUST00000203078.2
ENSMUST00000045235.8 |
Bmt2
|
base methyltransferase of 25S rRNA 2 |
| chr12_+_38830812 | 0.02 |
ENSMUST00000160856.8
|
Etv1
|
ets variant 1 |
| chr1_+_131898325 | 0.02 |
ENSMUST00000027695.8
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr8_+_94537910 | 0.02 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr15_+_78209920 | 0.02 |
ENSMUST00000230264.3
|
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr1_-_75119277 | 0.02 |
ENSMUST00000168720.8
ENSMUST00000041213.12 ENSMUST00000189809.2 |
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr3_-_30067537 | 0.02 |
ENSMUST00000108270.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr15_+_79231720 | 0.02 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr14_-_100522101 | 0.02 |
ENSMUST00000228216.2
|
Klf12
|
Kruppel-like factor 12 |
| chr11_-_99877423 | 0.02 |
ENSMUST00000105050.4
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr1_-_39760150 | 0.02 |
ENSMUST00000151913.3
|
Rfx8
|
regulatory factor X 8 |
| chr15_+_91722524 | 0.02 |
ENSMUST00000109276.8
ENSMUST00000088555.10 ENSMUST00000100293.9 ENSMUST00000126508.8 ENSMUST00000239545.1 |
Smgc
ENSMUSG00000118670.1
|
submandibular gland protein C mucin 19 |
| chr5_-_6926523 | 0.02 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr18_+_37827413 | 0.02 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr8_-_33374825 | 0.02 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1 |
| chr5_+_91051722 | 0.02 |
ENSMUST00000200681.4
ENSMUST00000075433.8 |
Cxcl2
|
chemokine (C-X-C motif) ligand 2 |
| chr7_+_44498415 | 0.02 |
ENSMUST00000107885.8
|
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr14_-_52150804 | 0.02 |
ENSMUST00000004673.15
ENSMUST00000111632.5 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr2_+_67004178 | 0.01 |
ENSMUST00000239009.2
ENSMUST00000238912.2 |
Xirp2
|
xin actin-binding repeat containing 2 |
| chr4_+_101365052 | 0.01 |
ENSMUST00000038207.12
|
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr2_-_86088672 | 0.01 |
ENSMUST00000216185.3
|
Olfr1049
|
olfactory receptor 1049 |
| chr14_-_52151026 | 0.01 |
ENSMUST00000228164.2
|
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr2_-_84652890 | 0.01 |
ENSMUST00000028471.6
|
Smtnl1
|
smoothelin-like 1 |
| chr15_-_58187556 | 0.01 |
ENSMUST00000022985.2
|
Klhl38
|
kelch-like 38 |
| chr14_+_50618620 | 0.01 |
ENSMUST00000215263.2
ENSMUST00000213402.2 ENSMUST00000213755.2 ENSMUST00000215227.2 |
Olfr736
|
olfactory receptor 736 |
| chr18_+_52958382 | 0.01 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr7_+_45433103 | 0.01 |
ENSMUST00000209617.2
ENSMUST00000209701.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr6_-_66656668 | 0.01 |
ENSMUST00000071414.2
|
Vmn1r35
|
vomeronasal 1 receptor 35 |
| chr11_+_118324652 | 0.01 |
ENSMUST00000106286.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_-_34083549 | 0.01 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr17_+_12597490 | 0.01 |
ENSMUST00000014578.7
|
Plg
|
plasminogen |
| chr6_-_57821483 | 0.01 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr10_-_41179167 | 0.01 |
ENSMUST00000043814.5
|
Fig4
|
FIG4 phosphoinositide 5-phosphatase |
| chr9_+_43983493 | 0.01 |
ENSMUST00000176671.2
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr15_-_78352801 | 0.01 |
ENSMUST00000229124.2
ENSMUST00000230226.2 ENSMUST00000017086.5 |
Tmprss6
|
transmembrane serine protease 6 |
| chr15_+_54274151 | 0.01 |
ENSMUST00000036737.4
|
Colec10
|
collectin sub-family member 10 |
| chr15_+_79232137 | 0.01 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr2_-_147888816 | 0.01 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr16_+_5703134 | 0.01 |
ENSMUST00000230658.2
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr15_-_66841465 | 0.01 |
ENSMUST00000170903.8
ENSMUST00000166420.8 ENSMUST00000005256.14 ENSMUST00000164070.2 |
Ndrg1
|
N-myc downstream regulated gene 1 |
| chr14_+_111912529 | 0.01 |
ENSMUST00000042767.9
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr2_-_86705146 | 0.01 |
ENSMUST00000130722.3
|
Olfr1096
|
olfactory receptor 1096 |
| chr11_+_69945157 | 0.01 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr8_-_33374282 | 0.01 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr7_+_45433306 | 0.01 |
ENSMUST00000072580.12
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr3_-_123483772 | 0.01 |
ENSMUST00000172537.3
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr17_+_70829050 | 0.01 |
ENSMUST00000133717.9
ENSMUST00000148486.8 |
Dlgap1
|
DLG associated protein 1 |
| chr3_-_106733603 | 0.01 |
ENSMUST00000213616.3
|
Olfr266
|
olfactory receptor 266 |
| chr9_-_99599312 | 0.01 |
ENSMUST00000112882.9
ENSMUST00000131922.2 |
Cldn18
|
claudin 18 |
| chr5_+_104473195 | 0.01 |
ENSMUST00000066207.4
|
Mepe
|
matrix extracellular phosphoglycoprotein with ASARM motif (bone) |
| chr7_-_12514534 | 0.01 |
ENSMUST00000211392.2
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr18_+_70058533 | 0.01 |
ENSMUST00000043929.11
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr1_+_83022653 | 0.01 |
ENSMUST00000222567.2
|
Gm47969
|
predicted gene, 47969 |
| chr3_-_30067285 | 0.01 |
ENSMUST00000172694.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr4_+_104623505 | 0.01 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chrX_+_162694397 | 0.01 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_+_68987257 | 0.01 |
ENSMUST00000167286.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_+_87501721 | 0.01 |
ENSMUST00000166259.8
ENSMUST00000172222.8 ENSMUST00000163606.8 |
Neu2
|
neuraminidase 2 |
| chr4_+_57821050 | 0.01 |
ENSMUST00000238994.2
|
Pakap
|
paralemmin A kinase anchor protein |
| chr18_+_70058613 | 0.01 |
ENSMUST00000080050.6
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr3_+_54063459 | 0.01 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr16_-_34083315 | 0.01 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr6_+_34840151 | 0.01 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr16_-_78887971 | 0.01 |
ENSMUST00000023566.11
ENSMUST00000060402.6 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr2_-_59955995 | 0.01 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr18_-_47466378 | 0.01 |
ENSMUST00000126684.2
ENSMUST00000156422.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_129811436 | 0.01 |
ENSMUST00000170920.2
|
Olfr247
|
olfactory receptor 247 |
| chr19_-_58443593 | 0.01 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr9_-_58026090 | 0.01 |
ENSMUST00000098681.4
ENSMUST00000098682.10 ENSMUST00000215944.2 |
Ccdc33
|
coiled-coil domain containing 33 |
| chr2_+_4722956 | 0.00 |
ENSMUST00000056914.7
|
Bend7
|
BEN domain containing 7 |
| chr4_+_135455427 | 0.00 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr8_+_59364789 | 0.00 |
ENSMUST00000062978.7
|
BC030500
|
cDNA sequence BC030500 |
| chr3_-_92922976 | 0.00 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr7_+_29991366 | 0.00 |
ENSMUST00000144508.2
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr12_-_111946560 | 0.00 |
ENSMUST00000190680.2
|
Rd3l
|
retinal degeneration 3-like |
| chr8_-_37420293 | 0.00 |
ENSMUST00000179501.2
|
Dlc1
|
deleted in liver cancer 1 |
| chr17_+_70829144 | 0.00 |
ENSMUST00000140728.8
|
Dlgap1
|
DLG associated protein 1 |
| chr15_+_91722458 | 0.00 |
ENSMUST00000109277.8
|
Smgc
|
submandibular gland protein C |
| chr3_-_144511566 | 0.00 |
ENSMUST00000199029.2
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr2_-_163486998 | 0.00 |
ENSMUST00000017851.4
|
Serinc3
|
serine incorporator 3 |
| chr11_-_68277799 | 0.00 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chrX_-_9983836 | 0.00 |
ENSMUST00000115543.3
ENSMUST00000044789.10 ENSMUST00000115544.9 |
Srpx
|
sushi-repeat-containing protein |
| chr10_+_115653152 | 0.00 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr7_+_90075762 | 0.00 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr10_-_41487315 | 0.00 |
ENSMUST00000219054.2
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr5_-_90788460 | 0.00 |
ENSMUST00000202704.4
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr14_+_58310143 | 0.00 |
ENSMUST00000022545.14
|
Fgf9
|
fibroblast growth factor 9 |
| chr1_-_87501548 | 0.00 |
ENSMUST00000068681.12
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr3_+_3699205 | 0.00 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr5_+_88073483 | 0.00 |
ENSMUST00000113271.3
|
Csn3
|
casein kappa |
| chr12_-_112477536 | 0.00 |
ENSMUST00000066791.7
|
Tmem179
|
transmembrane protein 179 |
| chr18_+_34891941 | 0.00 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr18_-_32271224 | 0.00 |
ENSMUST00000234657.2
ENSMUST00000234386.2 ENSMUST00000234651.2 |
Proc
|
protein C |
| chr1_-_180023467 | 0.00 |
ENSMUST00000161746.2
ENSMUST00000160879.7 |
Coq8a
|
coenzyme Q8A |
| chr7_+_141342696 | 0.00 |
ENSMUST00000155534.9
ENSMUST00000041924.14 ENSMUST00000163321.3 |
Muc5ac
|
mucin 5, subtypes A and C, tracheobronchial/gastric |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.4 | GO:2000096 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |