Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
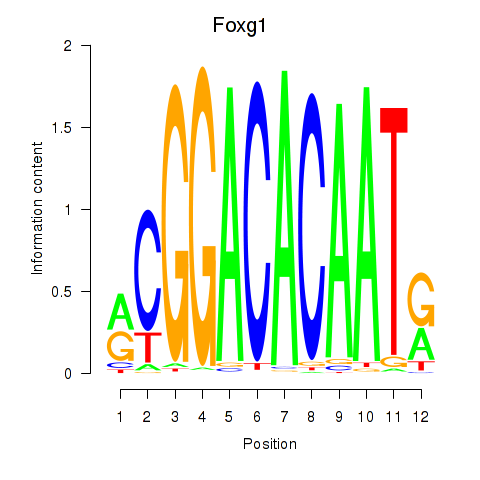
Results for Foxg1
Z-value: 0.77
Transcription factors associated with Foxg1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxg1
|
ENSMUSG00000020950.11 | forkhead box G1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxg1 | mm39_v1_chr12_+_49429574_49429666 | -0.60 | 2.9e-01 | Click! |
Activity profile of Foxg1 motif
Sorted Z-values of Foxg1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_34346936 | 0.58 |
ENSMUST00000124996.8
ENSMUST00000147632.3 |
Psmg4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr18_+_68433422 | 0.54 |
ENSMUST00000009679.11
ENSMUST00000131075.8 ENSMUST00000025427.14 ENSMUST00000139111.2 |
Rnmt
|
RNA (guanine-7-) methyltransferase |
| chr9_+_106048116 | 0.52 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chr3_-_50398027 | 0.41 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr15_+_79775819 | 0.37 |
ENSMUST00000177350.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr8_+_112263632 | 0.36 |
ENSMUST00000173506.8
|
Znrf1
|
zinc and ring finger 1 |
| chr5_-_121329385 | 0.31 |
ENSMUST00000054547.9
ENSMUST00000100770.9 |
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr3_-_106057077 | 0.24 |
ENSMUST00000149836.2
|
Chil3
|
chitinase-like 3 |
| chr1_-_180641099 | 0.23 |
ENSMUST00000159789.2
ENSMUST00000081026.11 |
H3f3a
|
H3.3 histone A |
| chr7_+_75498058 | 0.23 |
ENSMUST00000171155.4
ENSMUST00000092073.11 ENSMUST00000206019.2 ENSMUST00000205612.2 ENSMUST00000205887.2 |
Klhl25
|
kelch-like 25 |
| chr3_+_84081411 | 0.22 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr1_-_180641430 | 0.22 |
ENSMUST00000162814.8
|
H3f3a
|
H3.3 histone A |
| chr2_-_152672535 | 0.22 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr4_-_108690741 | 0.21 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr7_+_142558783 | 0.20 |
ENSMUST00000009396.13
|
Tspan32
|
tetraspanin 32 |
| chr1_-_180641159 | 0.20 |
ENSMUST00000162118.8
ENSMUST00000159685.2 ENSMUST00000161308.8 |
H3f3a
|
H3.3 histone A |
| chr2_-_154400753 | 0.20 |
ENSMUST00000109716.9
ENSMUST00000000895.13 ENSMUST00000125793.2 |
Necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr7_+_142558837 | 0.18 |
ENSMUST00000207211.2
|
Tspan32
|
tetraspanin 32 |
| chr19_+_11747721 | 0.18 |
ENSMUST00000167199.3
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr6_-_72322116 | 0.17 |
ENSMUST00000070345.5
|
Usp39
|
ubiquitin specific peptidase 39 |
| chr7_+_79848138 | 0.16 |
ENSMUST00000205822.2
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr18_-_68433398 | 0.16 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr5_+_145141333 | 0.15 |
ENSMUST00000085671.10
ENSMUST00000031601.8 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr9_+_54493784 | 0.15 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr15_+_79775894 | 0.14 |
ENSMUST00000177483.8
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr17_+_81251997 | 0.14 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr8_+_112263465 | 0.14 |
ENSMUST00000095176.12
|
Znrf1
|
zinc and ring finger 1 |
| chr14_-_121935829 | 0.13 |
ENSMUST00000040700.9
ENSMUST00000212181.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr2_+_28403255 | 0.12 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr7_-_41210219 | 0.12 |
ENSMUST00000164677.2
ENSMUST00000073410.12 |
Gm6871
|
predicted gene 6871 |
| chr11_-_97635484 | 0.12 |
ENSMUST00000018691.9
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr2_-_152673032 | 0.11 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr1_+_82817170 | 0.10 |
ENSMUST00000189220.7
ENSMUST00000113444.8 |
Agfg1
|
ArfGAP with FG repeats 1 |
| chr9_+_54493618 | 0.10 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr4_+_141520923 | 0.10 |
ENSMUST00000097805.11
ENSMUST00000030747.11 ENSMUST00000153094.2 |
Casp9
|
caspase 9 |
| chr11_-_119932573 | 0.09 |
ENSMUST00000103019.2
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr9_-_60557076 | 0.08 |
ENSMUST00000053171.14
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr8_+_112262729 | 0.08 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr12_-_112964279 | 0.07 |
ENSMUST00000011302.9
|
Brf1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr1_-_79838897 | 0.07 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr1_-_72914036 | 0.07 |
ENSMUST00000027377.9
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr8_+_93628015 | 0.07 |
ENSMUST00000104947.5
|
Capns2
|
calpain, small subunit 2 |
| chr2_+_28403084 | 0.05 |
ENSMUST00000135803.8
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr12_-_59058780 | 0.05 |
ENSMUST00000021375.12
|
Sec23a
|
SEC23 homolog A, COPII coat complex component |
| chr15_+_103148824 | 0.05 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr11_+_72498029 | 0.04 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr8_+_112263255 | 0.04 |
ENSMUST00000171182.8
ENSMUST00000168428.8 |
Znrf1
|
zinc and ring finger 1 |
| chr15_+_66449385 | 0.04 |
ENSMUST00000230882.2
ENSMUST00000048188.10 ENSMUST00000230948.2 ENSMUST00000229160.2 |
Phf20l1
|
PHD finger protein 20-like 1 |
| chr1_-_120432477 | 0.03 |
ENSMUST00000186432.3
|
Marco
|
macrophage receptor with collagenous structure |
| chr3_-_94922525 | 0.02 |
ENSMUST00000128438.2
ENSMUST00000149747.2 ENSMUST00000019482.8 |
Zfp687
|
zinc finger protein 687 |
| chr16_-_97306125 | 0.01 |
ENSMUST00000049721.9
ENSMUST00000231999.2 |
Fam3b
|
family with sequence similarity 3, member B |
| chr11_-_99684173 | 0.01 |
ENSMUST00000105058.2
|
Gm11596
|
predicted gene 11596 |
| chr2_-_89159994 | 0.01 |
ENSMUST00000213860.2
ENSMUST00000215679.2 |
Olfr1232
|
olfactory receptor 1232 |
| chr1_+_180762587 | 0.01 |
ENSMUST00000037361.9
|
Lefty1
|
left right determination factor 1 |
| chr2_-_162929732 | 0.01 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr16_-_28748410 | 0.01 |
ENSMUST00000100023.3
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr6_-_128503666 | 0.01 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr7_-_21053786 | 0.00 |
ENSMUST00000105199.2
|
Vmn1r127
|
vomeronasal 1 receptor 127 |
| chr2_+_74522258 | 0.00 |
ENSMUST00000061745.5
|
Hoxd10
|
homeobox D10 |
| chr7_-_21496153 | 0.00 |
ENSMUST00000105196.2
|
Vmn1r248
|
vomeronasal 1 receptor 248 |
| chr7_-_22719348 | 0.00 |
ENSMUST00000168926.2
|
Vmn1r259
|
vomeronasal 1 receptor 259 |
| chr2_+_120331784 | 0.00 |
ENSMUST00000151342.3
|
Capn3
|
calpain 3 |
| chr13_+_21887145 | 0.00 |
ENSMUST00000077843.6
|
Olfr1359
|
olfactory receptor 1359 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxg1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.3 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.5 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.2 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.4 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |