Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
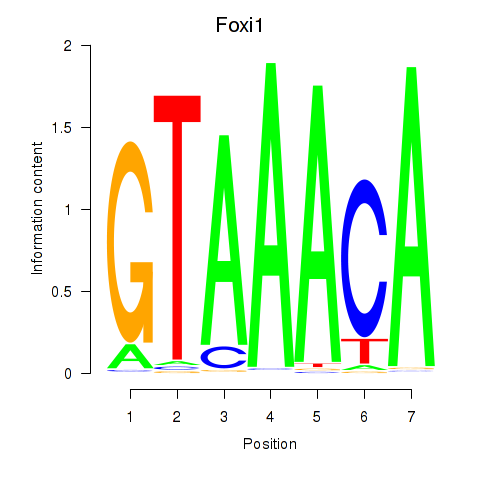
Results for Foxi1_Foxo1
Z-value: 0.56
Transcription factors associated with Foxi1_Foxo1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxi1
|
ENSMUSG00000047861.3 | forkhead box I1 |
|
Foxo1
|
ENSMUSG00000044167.7 | forkhead box O1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxi1 | mm39_v1_chr11_-_34158085_34158092 | -0.62 | 2.6e-01 | Click! |
| Foxo1 | mm39_v1_chr3_+_52175757_52175827 | -0.39 | 5.1e-01 | Click! |
Activity profile of Foxi1_Foxo1 motif
Sorted Z-values of Foxi1_Foxo1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_138169253 | 0.65 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_138169509 | 0.55 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr11_+_3280401 | 0.38 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr17_+_47816137 | 0.34 |
ENSMUST00000182935.8
ENSMUST00000182506.8 |
Ccnd3
|
cyclin D3 |
| chr1_+_40554513 | 0.33 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr14_-_31503869 | 0.31 |
ENSMUST00000227089.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr11_+_3280771 | 0.30 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chrX_+_168468186 | 0.30 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr8_+_106785434 | 0.30 |
ENSMUST00000212742.2
ENSMUST00000211991.2 |
Nfatc3
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 3 |
| chr3_-_146475974 | 0.27 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr17_+_29309942 | 0.27 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr11_-_102255999 | 0.26 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr1_+_79753735 | 0.26 |
ENSMUST00000027464.9
|
Mrpl44
|
mitochondrial ribosomal protein L44 |
| chr12_-_71183371 | 0.26 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr5_-_138169476 | 0.25 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr4_-_87724533 | 0.24 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr16_+_16964801 | 0.24 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chr16_-_92196954 | 0.23 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr12_+_52144511 | 0.23 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr17_+_47816074 | 0.23 |
ENSMUST00000183177.8
ENSMUST00000182848.8 |
Ccnd3
|
cyclin D3 |
| chr10_-_93146825 | 0.22 |
ENSMUST00000151153.2
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr1_-_171108754 | 0.21 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr19_+_46611826 | 0.20 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr5_-_92496730 | 0.20 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr3_+_102965910 | 0.20 |
ENSMUST00000199367.5
ENSMUST00000199049.5 ENSMUST00000197678.5 |
Nras
|
neuroblastoma ras oncogene |
| chr5_-_115236354 | 0.20 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr7_+_141056305 | 0.20 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr6_-_124888643 | 0.20 |
ENSMUST00000032217.2
|
Lag3
|
lymphocyte-activation gene 3 |
| chr19_-_41836514 | 0.20 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr3_+_106020545 | 0.19 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr19_-_8691460 | 0.19 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr18_+_13107535 | 0.19 |
ENSMUST00000234035.2
ENSMUST00000235053.2 |
Impact
|
impact, RWD domain protein |
| chr12_+_104202493 | 0.19 |
ENSMUST00000043315.15
ENSMUST00000171916.8 ENSMUST00000170628.2 |
Serpina3g
|
serine (or cysteine) peptidase inhibitor, clade A, member 3G |
| chr7_+_115692530 | 0.19 |
ENSMUST00000032899.12
ENSMUST00000106608.8 ENSMUST00000106607.2 |
1110004F10Rik
|
RIKEN cDNA 1110004F10 gene |
| chr11_+_120123727 | 0.19 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr4_-_15945359 | 0.19 |
ENSMUST00000029877.9
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr19_+_45433899 | 0.19 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr2_+_173579285 | 0.18 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr2_-_91274967 | 0.18 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr7_-_103463120 | 0.18 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr17_-_27816151 | 0.18 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr10_-_37014859 | 0.18 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr17_+_47815968 | 0.17 |
ENSMUST00000182129.8
ENSMUST00000171031.8 |
Ccnd3
|
cyclin D3 |
| chr7_-_25488060 | 0.17 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr11_+_3282424 | 0.17 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr10_-_79911245 | 0.17 |
ENSMUST00000217972.2
|
Sbno2
|
strawberry notch 2 |
| chr19_+_7534838 | 0.17 |
ENSMUST00000141887.8
ENSMUST00000136756.2 |
Plaat3
|
phospholipase A and acyltransferase 3 |
| chr13_+_4241149 | 0.17 |
ENSMUST00000021634.4
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr4_+_108704982 | 0.17 |
ENSMUST00000102738.4
|
Kti12
|
KTI12 homolog, chromatin associated |
| chr5_-_137530214 | 0.17 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr17_+_34251322 | 0.16 |
ENSMUST00000116612.3
|
Rxrb
|
retinoid X receptor beta |
| chrX_+_100298134 | 0.16 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr5_-_134205559 | 0.16 |
ENSMUST00000076228.3
|
Rcc1l
|
reculator of chromosome condensation 1 like |
| chr1_+_52158721 | 0.16 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr5_-_124717146 | 0.16 |
ENSMUST00000031334.15
|
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chr17_+_41121979 | 0.16 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr13_-_98152768 | 0.16 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr17_+_85265420 | 0.16 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr3_+_85946145 | 0.16 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr14_+_54496683 | 0.16 |
ENSMUST00000041197.13
ENSMUST00000239403.2 ENSMUST00000197605.3 |
Abhd4
|
abhydrolase domain containing 4 |
| chr11_+_98632696 | 0.16 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr3_+_138566249 | 0.16 |
ENSMUST00000121826.3
|
Tspan5
|
tetraspanin 5 |
| chr3_-_27207931 | 0.15 |
ENSMUST00000175857.2
ENSMUST00000177055.8 ENSMUST00000176535.8 |
Ect2
|
ect2 oncogene |
| chr5_-_124717055 | 0.15 |
ENSMUST00000128920.2
ENSMUST00000135361.8 |
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chr2_+_28531239 | 0.15 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr15_-_38518458 | 0.15 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr18_+_36431732 | 0.15 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr9_-_45866264 | 0.15 |
ENSMUST00000114573.9
|
Sidt2
|
SID1 transmembrane family, member 2 |
| chr14_-_54651442 | 0.15 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr10_+_79951613 | 0.15 |
ENSMUST00000003152.14
|
Stk11
|
serine/threonine kinase 11 |
| chrX_+_7588505 | 0.15 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr6_-_83808717 | 0.15 |
ENSMUST00000058383.9
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr4_-_129534403 | 0.15 |
ENSMUST00000084264.12
|
Txlna
|
taxilin alpha |
| chr8_+_47193275 | 0.15 |
ENSMUST00000207571.3
|
Irf2
|
interferon regulatory factor 2 |
| chr4_-_86587728 | 0.14 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr2_+_4887015 | 0.14 |
ENSMUST00000115019.2
|
Sephs1
|
selenophosphate synthetase 1 |
| chr15_-_103218876 | 0.14 |
ENSMUST00000079824.6
|
Gpr84
|
G protein-coupled receptor 84 |
| chr15_-_98816012 | 0.14 |
ENSMUST00000023736.10
|
Lmbr1l
|
limb region 1 like |
| chr12_+_59060162 | 0.14 |
ENSMUST00000021379.8
|
Gemin2
|
gem nuclear organelle associated protein 2 |
| chr17_+_34124078 | 0.14 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr7_-_25089522 | 0.14 |
ENSMUST00000054301.14
|
Lipe
|
lipase, hormone sensitive |
| chr5_-_105441554 | 0.14 |
ENSMUST00000050011.10
ENSMUST00000196520.2 |
Gm43302
Gbp6
|
predicted gene 43302 guanylate binding protein 6 |
| chr3_-_90603013 | 0.14 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr17_+_47816042 | 0.14 |
ENSMUST00000183044.8
ENSMUST00000037333.17 |
Ccnd3
|
cyclin D3 |
| chr5_-_137529251 | 0.14 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr8_-_25215778 | 0.14 |
ENSMUST00000171438.8
ENSMUST00000171611.9 |
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chr3_+_129326004 | 0.14 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr5_+_21577640 | 0.14 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr1_+_52158599 | 0.14 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr5_-_137529465 | 0.14 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr19_+_56414114 | 0.14 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr4_+_102617332 | 0.13 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_+_144619696 | 0.13 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr12_+_103400470 | 0.13 |
ENSMUST00000079294.12
ENSMUST00000076788.12 ENSMUST00000076702.12 ENSMUST00000066701.13 ENSMUST00000085065.12 ENSMUST00000140838.2 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr11_-_59727752 | 0.13 |
ENSMUST00000156837.2
|
Cops3
|
COP9 signalosome subunit 3 |
| chr18_+_69477541 | 0.13 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr14_+_55798362 | 0.13 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr2_+_38401826 | 0.13 |
ENSMUST00000112895.8
|
Nek6
|
NIMA (never in mitosis gene a)-related expressed kinase 6 |
| chr19_-_60770628 | 0.13 |
ENSMUST00000238125.2
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr13_-_53083494 | 0.13 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr13_+_104315301 | 0.13 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chrX_-_98514278 | 0.13 |
ENSMUST00000113797.4
ENSMUST00000113790.8 ENSMUST00000036354.7 ENSMUST00000167246.2 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr17_+_29712008 | 0.13 |
ENSMUST00000234665.2
|
Pim1
|
proviral integration site 1 |
| chr7_+_125151292 | 0.13 |
ENSMUST00000033004.8
|
Il4ra
|
interleukin 4 receptor, alpha |
| chr5_+_110478558 | 0.13 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr4_-_149760488 | 0.13 |
ENSMUST00000118704.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr10_-_126866682 | 0.13 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr1_-_144124871 | 0.13 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr3_+_51568625 | 0.13 |
ENSMUST00000159554.7
ENSMUST00000161590.4 |
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr16_-_22258469 | 0.13 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr13_+_94954202 | 0.13 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr5_-_65090893 | 0.13 |
ENSMUST00000197315.5
|
Tlr1
|
toll-like receptor 1 |
| chr19_-_45771939 | 0.12 |
ENSMUST00000026243.5
|
Oga
|
O-GlcNAcase |
| chr19_+_36325683 | 0.12 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr3_-_27207993 | 0.12 |
ENSMUST00000176242.9
ENSMUST00000176780.8 |
Ect2
|
ect2 oncogene |
| chr6_+_41523664 | 0.12 |
ENSMUST00000103299.3
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr1_+_52158693 | 0.12 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_+_63769772 | 0.12 |
ENSMUST00000027103.7
|
Fastkd2
|
FAST kinase domains 2 |
| chr9_-_100388857 | 0.12 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr10_-_88601443 | 0.12 |
ENSMUST00000220188.2
ENSMUST00000218967.2 |
Utp20
|
UTP20 small subunit processome component |
| chr6_-_71417607 | 0.12 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr8_-_25215856 | 0.12 |
ENSMUST00000033958.15
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin) |
| chrX_+_149330371 | 0.12 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr11_-_3216377 | 0.12 |
ENSMUST00000020741.12
|
Drg1
|
developmentally regulated GTP binding protein 1 |
| chr13_+_21365308 | 0.12 |
ENSMUST00000221464.2
|
Trim27
|
tripartite motif-containing 27 |
| chr5_+_148239975 | 0.12 |
ENSMUST00000152105.8
ENSMUST00000085554.5 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr2_+_121859025 | 0.12 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr6_+_134807097 | 0.12 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr5_+_139239247 | 0.12 |
ENSMUST00000138508.8
ENSMUST00000110878.2 |
Get4
|
golgi to ER traffic protein 4 |
| chr7_+_125202653 | 0.12 |
ENSMUST00000206103.2
ENSMUST00000033000.8 |
Il21r
|
interleukin 21 receptor |
| chr10_-_80513927 | 0.12 |
ENSMUST00000199949.2
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr13_-_63712349 | 0.11 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr7_+_125151432 | 0.11 |
ENSMUST00000206217.2
ENSMUST00000205985.2 |
Il4ra
|
interleukin 4 receptor, alpha |
| chr11_-_20691440 | 0.11 |
ENSMUST00000177543.8
|
Aftph
|
aftiphilin |
| chr15_-_60696790 | 0.11 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr6_+_52691204 | 0.11 |
ENSMUST00000138040.8
ENSMUST00000129660.2 |
Tax1bp1
|
Tax1 (human T cell leukemia virus type I) binding protein 1 |
| chr18_-_39622829 | 0.11 |
ENSMUST00000025300.13
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr12_-_69771604 | 0.11 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr6_-_100648086 | 0.11 |
ENSMUST00000089245.7
ENSMUST00000113312.9 ENSMUST00000170667.8 |
Shq1
|
SHQ1 homolog (S. cerevisiae) |
| chr1_-_45964730 | 0.11 |
ENSMUST00000027137.11
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr2_-_106800736 | 0.11 |
ENSMUST00000093883.4
|
Arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr11_-_59700820 | 0.11 |
ENSMUST00000047706.3
ENSMUST00000102697.10 |
Flcn
|
folliculin |
| chr6_-_7693184 | 0.11 |
ENSMUST00000031766.12
|
Asns
|
asparagine synthetase |
| chr10_-_128384971 | 0.11 |
ENSMUST00000176906.2
|
Rpl41
|
ribosomal protein L41 |
| chr17_-_71158703 | 0.11 |
ENSMUST00000166395.9
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr4_-_117744476 | 0.11 |
ENSMUST00000132073.2
ENSMUST00000150204.8 ENSMUST00000147845.2 ENSMUST00000036380.14 ENSMUST00000136596.2 |
Atp6v0b
|
ATPase, H+ transporting, lysosomal V0 subunit B |
| chr14_-_31362835 | 0.11 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr12_-_83643883 | 0.11 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr14_+_55798517 | 0.11 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chrX_-_139443926 | 0.11 |
ENSMUST00000055738.12
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr10_-_93727003 | 0.11 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr7_-_133384449 | 0.11 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr1_-_45542442 | 0.11 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr7_+_27290969 | 0.11 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr13_+_55593116 | 0.11 |
ENSMUST00000001115.16
ENSMUST00000224995.2 ENSMUST00000225925.2 ENSMUST00000099482.5 ENSMUST00000224118.2 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr4_-_42856771 | 0.10 |
ENSMUST00000107981.2
|
Gm12394
|
predicted gene 12394 |
| chr1_-_192923816 | 0.10 |
ENSMUST00000160929.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr2_-_32594156 | 0.10 |
ENSMUST00000127812.3
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr17_+_26895344 | 0.10 |
ENSMUST00000015719.16
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr7_-_125976580 | 0.10 |
ENSMUST00000119846.8
ENSMUST00000119754.8 ENSMUST00000032994.15 |
Spns1
|
spinster homolog 1 |
| chr19_+_29078765 | 0.10 |
ENSMUST00000064393.6
ENSMUST00000235900.2 |
Rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr5_+_124717197 | 0.10 |
ENSMUST00000031333.4
|
Gtf2h3
|
general transcription factor IIH, polypeptide 3 |
| chr10_-_112764879 | 0.10 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr7_+_90091937 | 0.10 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr10_+_11157326 | 0.10 |
ENSMUST00000070300.5
|
Fbxo30
|
F-box protein 30 |
| chr10_+_116013256 | 0.10 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr10_+_96453408 | 0.10 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr14_-_71003973 | 0.10 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr11_+_98632953 | 0.10 |
ENSMUST00000153043.8
|
Thra
|
thyroid hormone receptor alpha |
| chr13_+_28441511 | 0.10 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr19_-_32717166 | 0.10 |
ENSMUST00000235142.2
ENSMUST00000070210.6 ENSMUST00000236011.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr4_-_129556234 | 0.10 |
ENSMUST00000003828.11
|
Kpna6
|
karyopherin (importin) alpha 6 |
| chr5_+_122347912 | 0.10 |
ENSMUST00000143560.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr18_-_43610829 | 0.10 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr8_-_79539838 | 0.10 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr3_+_51568588 | 0.10 |
ENSMUST00000099106.10
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr1_-_16727133 | 0.10 |
ENSMUST00000185771.7
|
Eloc
|
elongin C |
| chr3_+_129326285 | 0.10 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr19_-_38032006 | 0.10 |
ENSMUST00000172095.3
ENSMUST00000041475.16 |
Myof
|
myoferlin |
| chr1_+_91249797 | 0.10 |
ENSMUST00000088904.10
|
Espnl
|
espin-like |
| chr1_-_185061525 | 0.10 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr18_+_34891941 | 0.10 |
ENSMUST00000049281.12
|
Fam53c
|
family with sequence similarity 53, member C |
| chr15_-_36496880 | 0.10 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr9_+_108437485 | 0.10 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr9_-_14292453 | 0.10 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr11_-_115915315 | 0.10 |
ENSMUST00000016703.8
|
H3f3b
|
H3.3 histone B |
| chr7_-_68398989 | 0.10 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr14_-_56322654 | 0.10 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr6_+_29853745 | 0.10 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chrX_+_138464065 | 0.10 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr12_-_108241597 | 0.10 |
ENSMUST00000222310.2
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr2_-_101451383 | 0.10 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr17_+_26342474 | 0.10 |
ENSMUST00000025014.10
ENSMUST00000236166.2 ENSMUST00000127647.3 |
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr2_-_71198091 | 0.10 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr2_+_122461079 | 0.10 |
ENSMUST00000239506.1
|
SPATA5L1
|
spermatosis associated 5-like 1 |
| chr11_-_48836975 | 0.09 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr18_+_47245204 | 0.09 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr10_-_116899664 | 0.09 |
ENSMUST00000218719.2
ENSMUST00000219573.2 ENSMUST00000047672.9 |
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr11_+_80320558 | 0.09 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr11_-_40586029 | 0.09 |
ENSMUST00000101347.10
|
Mat2b
|
methionine adenosyltransferase II, beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxi1_Foxo1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.1 | 1.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.9 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:0072755 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.1 | 0.2 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.1 | 0.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.1 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.2 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.2 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.0 | 0.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:1901873 | negative regulation of cellular respiration(GO:1901856) regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:1902267 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.2 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.5 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.0 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.0 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:1900864 | tRNA wobble position uridine thiolation(GO:0002143) mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.0 | 0.0 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.0 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.0 | 0.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0001762 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.0 | 0.0 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.0 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.0 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.0 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.0 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.0 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:1900245 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0030681 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.0 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.2 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.1 | 0.3 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.2 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.3 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.3 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.0 | GO:0047223 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.0 | GO:0004844 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.2 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.1 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.0 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |