Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
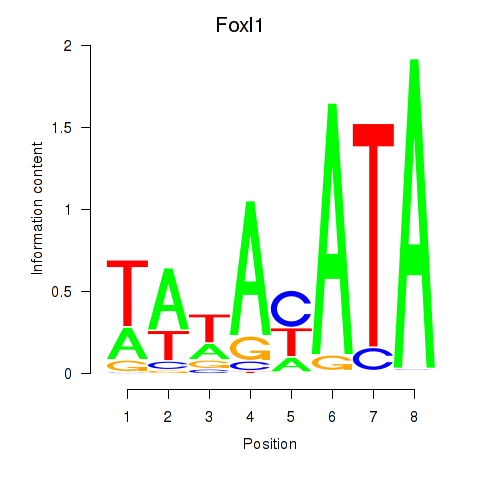
Results for Foxl1
Z-value: 1.08
Transcription factors associated with Foxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxl1
|
ENSMUSG00000097084.2 | forkhead box L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxl1 | mm39_v1_chr8_+_121854566_121854679 | 0.68 | 2.0e-01 | Click! |
Activity profile of Foxl1 motif
Sorted Z-values of Foxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23947641 | 2.79 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr13_-_23806530 | 2.65 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr13_+_23922783 | 1.35 |
ENSMUST00000040914.3
|
H1f2
|
H1.2 linker histone, cluster member |
| chrM_+_2743 | 0.61 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr10_-_127190280 | 0.61 |
ENSMUST00000059718.6
|
Inhbe
|
inhibin beta-E |
| chr11_+_31822211 | 0.37 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_+_18059982 | 0.31 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr4_-_120874376 | 0.29 |
ENSMUST00000043200.8
|
Smap2
|
small ArfGAP 2 |
| chr3_+_144283355 | 0.27 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F |
| chr10_+_56253418 | 0.23 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr16_+_44913974 | 0.22 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr15_-_11905697 | 0.20 |
ENSMUST00000066529.5
ENSMUST00000228603.2 |
Npr3
|
natriuretic peptide receptor 3 |
| chr5_+_102629240 | 0.16 |
ENSMUST00000073302.12
ENSMUST00000094559.9 |
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_+_102629365 | 0.14 |
ENSMUST00000112854.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr9_-_101076198 | 0.11 |
ENSMUST00000066773.9
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_-_169938060 | 0.10 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr6_-_41751648 | 0.07 |
ENSMUST00000214752.2
|
Olfr459
|
olfactory receptor 459 |
| chr6_-_102441628 | 0.07 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr18_-_15284476 | 0.05 |
ENSMUST00000025992.7
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr4_+_11191354 | 0.03 |
ENSMUST00000170901.8
|
Ccne2
|
cyclin E2 |
| chr10_+_20046748 | 0.01 |
ENSMUST00000215924.2
|
Map7
|
microtubule-associated protein 7 |
| chr2_+_85822163 | 0.01 |
ENSMUST00000050942.3
|
Olfr1031
|
olfactory receptor 1031 |
| chr18_+_37637317 | 0.00 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chrX_+_111513971 | 0.00 |
ENSMUST00000071814.13
|
Zfp711
|
zinc finger protein 711 |
| chr2_+_91086299 | 0.00 |
ENSMUST00000134699.8
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.6 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0005922 | fascia adherens(GO:0005916) connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |