Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
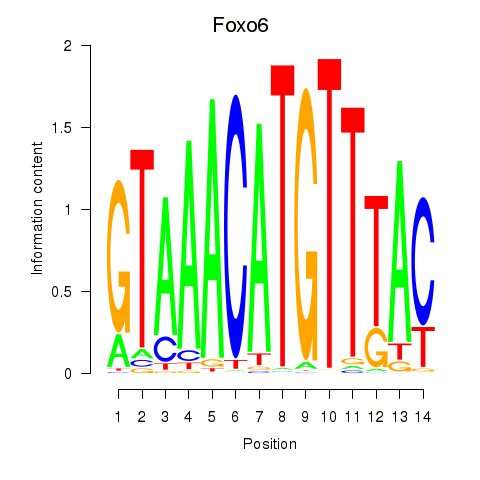
Results for Foxo6
Z-value: 0.24
Transcription factors associated with Foxo6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo6
|
ENSMUSG00000052135.9 | forkhead box O6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo6 | mm39_v1_chr4_-_120144546_120144546 | -0.97 | 6.5e-03 | Click! |
Activity profile of Foxo6 motif
Sorted Z-values of Foxo6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87684299 | 0.25 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr19_+_8870362 | 0.20 |
ENSMUST00000096249.7
|
Ints5
|
integrator complex subunit 5 |
| chr11_-_59727752 | 0.19 |
ENSMUST00000156837.2
|
Cops3
|
COP9 signalosome subunit 3 |
| chr4_-_136626073 | 0.18 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr13_-_96803216 | 0.16 |
ENSMUST00000170287.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr15_+_103362195 | 0.15 |
ENSMUST00000047405.9
|
Nckap1l
|
NCK associated protein 1 like |
| chr16_+_16964801 | 0.13 |
ENSMUST00000232479.2
ENSMUST00000232344.2 ENSMUST00000069064.7 |
Ydjc
|
YdjC homolog (bacterial) |
| chr5_+_110478558 | 0.11 |
ENSMUST00000112481.2
|
Pole
|
polymerase (DNA directed), epsilon |
| chr7_+_142606476 | 0.11 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr3_+_103875261 | 0.11 |
ENSMUST00000117150.8
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr5_-_117527094 | 0.11 |
ENSMUST00000111953.2
ENSMUST00000086461.13 |
Rfc5
|
replication factor C (activator 1) 5 |
| chr3_+_103875574 | 0.11 |
ENSMUST00000063717.14
ENSMUST00000055425.15 ENSMUST00000123611.8 ENSMUST00000090685.11 |
Phtf1
|
putative homeodomain transcription factor 1 |
| chr4_+_134238310 | 0.11 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr17_-_33879224 | 0.10 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chrX_-_8059597 | 0.10 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr6_-_129449739 | 0.10 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr2_-_92876398 | 0.10 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chr17_+_64203017 | 0.09 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr6_+_59185846 | 0.09 |
ENSMUST00000062626.4
|
Tigd2
|
tigger transposable element derived 2 |
| chr6_+_57679621 | 0.09 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr17_-_30831576 | 0.08 |
ENSMUST00000235171.2
ENSMUST00000236335.2 ENSMUST00000167624.2 |
Glo1
|
glyoxalase 1 |
| chr4_+_102427247 | 0.08 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr17_-_23783063 | 0.08 |
ENSMUST00000095606.5
|
Zfp213
|
zinc finger protein 213 |
| chr14_-_70945434 | 0.08 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr1_-_180073492 | 0.07 |
ENSMUST00000010753.14
|
Psen2
|
presenilin 2 |
| chr2_-_25498459 | 0.07 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr1_-_180073322 | 0.07 |
ENSMUST00000111104.2
|
Psen2
|
presenilin 2 |
| chr18_-_32044877 | 0.07 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr3_-_127574167 | 0.07 |
ENSMUST00000029662.12
|
Alpk1
|
alpha-kinase 1 |
| chr6_+_57679455 | 0.07 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr11_+_49916136 | 0.07 |
ENSMUST00000054684.14
ENSMUST00000238748.2 ENSMUST00000102776.5 |
Rnf130
|
ring finger protein 130 |
| chr5_+_115417725 | 0.06 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr17_+_48653493 | 0.06 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr15_+_41573995 | 0.06 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr4_+_108336164 | 0.06 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr18_+_60345152 | 0.06 |
ENSMUST00000031549.6
|
Gm4951
|
predicted gene 4951 |
| chr2_-_84255602 | 0.06 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr13_+_76727787 | 0.06 |
ENSMUST00000126960.8
ENSMUST00000109583.9 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr9_+_7272514 | 0.06 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr2_-_155199300 | 0.06 |
ENSMUST00000165234.2
ENSMUST00000077626.13 |
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr15_+_75140241 | 0.06 |
ENSMUST00000023247.13
|
Ly6f
|
lymphocyte antigen 6 complex, locus F |
| chr18_-_12254562 | 0.05 |
ENSMUST00000055447.14
ENSMUST00000209859.2 |
Tmem241
|
transmembrane protein 241 |
| chr5_-_88823049 | 0.05 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr2_-_30084124 | 0.05 |
ENSMUST00000113659.8
ENSMUST00000113660.2 |
Kyat1
|
kynurenine aminotransferase 1 |
| chrX_-_73325204 | 0.05 |
ENSMUST00000114189.10
ENSMUST00000119361.4 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr15_-_74855264 | 0.05 |
ENSMUST00000023250.11
ENSMUST00000166694.2 |
Ly6i
|
lymphocyte antigen 6 complex, locus I |
| chr1_-_65158717 | 0.05 |
ENSMUST00000144760.3
|
Gm28845
|
predicted gene 28845 |
| chr13_-_4329421 | 0.04 |
ENSMUST00000021632.5
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr2_+_118428690 | 0.04 |
ENSMUST00000038341.8
|
Bub1b
|
BUB1B, mitotic checkpoint serine/threonine kinase |
| chr2_+_120807498 | 0.04 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr5_-_65548723 | 0.04 |
ENSMUST00000120094.5
ENSMUST00000127874.6 ENSMUST00000057885.13 |
Rpl9
|
ribosomal protein L9 |
| chr12_-_102390000 | 0.04 |
ENSMUST00000110020.8
|
Lgmn
|
legumain |
| chr14_+_31888061 | 0.04 |
ENSMUST00000164341.2
|
Ncoa4
|
nuclear receptor coactivator 4 |
| chr5_+_65548840 | 0.04 |
ENSMUST00000122026.6
ENSMUST00000031101.10 ENSMUST00000200374.2 |
Lias
|
lipoic acid synthetase |
| chr5_-_52347826 | 0.04 |
ENSMUST00000199321.5
ENSMUST00000195922.2 ENSMUST00000031061.12 |
Dhx15
|
DEAH (Asp-Glu-Ala-His) box polypeptide 15 |
| chr15_+_52575796 | 0.03 |
ENSMUST00000037115.9
|
Med30
|
mediator complex subunit 30 |
| chr1_+_179788675 | 0.03 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr10_+_29189159 | 0.03 |
ENSMUST00000160399.8
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr18_+_37884653 | 0.03 |
ENSMUST00000194928.2
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr15_+_100052260 | 0.02 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr2_+_163916042 | 0.02 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr10_+_39296005 | 0.02 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr11_+_87744033 | 0.02 |
ENSMUST00000038196.7
|
Mks1
|
MKS transition zone complex subunit 1 |
| chr12_-_113802603 | 0.02 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr7_+_28508220 | 0.02 |
ENSMUST00000172529.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr6_-_54949587 | 0.02 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr4_-_62389098 | 0.02 |
ENSMUST00000135811.2
ENSMUST00000120095.8 ENSMUST00000030087.14 ENSMUST00000107452.8 ENSMUST00000155522.8 |
Wdr31
|
WD repeat domain 31 |
| chr7_+_122969004 | 0.01 |
ENSMUST00000206721.2
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr5_+_108777855 | 0.01 |
ENSMUST00000078323.13
ENSMUST00000120327.3 |
Tmem175
|
transmembrane protein 175 |
| chr2_-_130928658 | 0.01 |
ENSMUST00000028794.10
ENSMUST00000110227.8 ENSMUST00000110226.2 |
Siglec1
|
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr6_-_48063464 | 0.01 |
ENSMUST00000073124.9
ENSMUST00000203609.3 |
Zfp746
|
zinc finger protein 746 |
| chr18_+_5593566 | 0.01 |
ENSMUST00000160910.2
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr17_-_31496301 | 0.01 |
ENSMUST00000235144.2
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr7_+_28140450 | 0.01 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma |
| chr10_-_130333265 | 0.01 |
ENSMUST00000164227.4
|
Vmn2r87
|
vomeronasal 2, receptor 87 |
| chr9_+_19031678 | 0.01 |
ENSMUST00000212730.3
|
Olfr836
|
olfactory receptor 836 |
| chr3_-_51316347 | 0.01 |
ENSMUST00000193279.2
ENSMUST00000038108.12 |
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr9_-_51240201 | 0.01 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr16_-_93382146 | 0.01 |
ENSMUST00000232835.2
|
Setd4
|
SET domain containing 4 |
| chr9_-_21982637 | 0.01 |
ENSMUST00000179605.9
ENSMUST00000043922.7 |
Zfp653
|
zinc finger protein 653 |
| chr14_-_49482846 | 0.01 |
ENSMUST00000227113.2
ENSMUST00000130853.2 ENSMUST00000228936.2 ENSMUST00000022398.15 |
ccdc198
|
coiled-coil domain containing 198 |
| chr16_-_59151501 | 0.01 |
ENSMUST00000213910.2
|
Olfr205
|
olfactory receptor 205 |
| chr11_+_99676017 | 0.01 |
ENSMUST00000105059.4
|
Krtap4-9
|
keratin associated protein 4-9 |
| chr3_-_49711765 | 0.01 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr3_-_49711706 | 0.01 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr5_-_87240405 | 0.01 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr6_-_57821483 | 0.01 |
ENSMUST00000226191.2
|
Vmn1r21
|
vomeronasal 1 receptor 21 |
| chr14_+_51648458 | 0.01 |
ENSMUST00000022438.12
|
Vmn2r88
|
vomeronasal 2, receptor 88 |
| chr6_+_40546421 | 0.01 |
ENSMUST00000216942.2
|
Olfr460
|
olfactory receptor 460 |
| chr2_-_89406117 | 0.01 |
ENSMUST00000214870.2
|
Olfr1245
|
olfactory receptor 1245 |
| chr16_+_84312257 | 0.01 |
ENSMUST00000210306.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr5_-_87288177 | 0.01 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr11_-_11977958 | 0.01 |
ENSMUST00000109654.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr6_-_68784692 | 0.01 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr14_+_96118660 | 0.01 |
ENSMUST00000228913.2
ENSMUST00000045892.3 |
Spertl
|
spermatid associated like |
| chr7_-_19902302 | 0.00 |
ENSMUST00000164526.2
|
Vmn1r94
|
vomeronasal 1 receptor 94 |
| chr14_+_53921052 | 0.00 |
ENSMUST00000103663.6
|
Trav4-4-dv10
|
T cell receptor alpha variable 4-4-DV10 |
| chr16_-_59182579 | 0.00 |
ENSMUST00000055868.4
ENSMUST00000208246.2 |
Olfr209
|
olfactory receptor 209 |
| chrX_+_56454507 | 0.00 |
ENSMUST00000114725.3
|
Tm9sf5
|
transmembrane 9 superfamily member 5 |
| chr5_+_87148697 | 0.00 |
ENSMUST00000031186.9
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr5_-_86780277 | 0.00 |
ENSMUST00000116553.9
|
Tmprss11f
|
transmembrane protease, serine 11f |
| chr7_+_29883569 | 0.00 |
ENSMUST00000098594.4
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr2_-_120946877 | 0.00 |
ENSMUST00000110675.3
|
Tgm7
|
transglutaminase 7 |
| chr2_+_87039314 | 0.00 |
ENSMUST00000215611.2
|
Olfr1113
|
olfactory receptor 1113 |
| chr7_-_104542833 | 0.00 |
ENSMUST00000081116.2
|
Olfr666
|
olfactory receptor 666 |
| chr7_-_9687512 | 0.00 |
ENSMUST00000165611.2
|
Vmn2r48
|
vomeronasal 2, receptor 48 |
| chrX_+_143253677 | 0.00 |
ENSMUST00000178233.2
|
Trpc5os
|
transient receptor potential cation channel, subfamily C, member 5, opposite strand |
| chr1_+_100025486 | 0.00 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr2_+_36796923 | 0.00 |
ENSMUST00000068475.5
|
Olfr354
|
olfactory receptor 354 |
| chr5_+_92534738 | 0.00 |
ENSMUST00000128246.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr2_+_86616076 | 0.00 |
ENSMUST00000055273.2
|
Olfr1093
|
olfactory receptor 1093 |
| chr10_+_33913593 | 0.00 |
ENSMUST00000095758.3
|
Trappc3l
|
trafficking protein particle complex 3 like |
| chr14_+_53478202 | 0.00 |
ENSMUST00000179583.3
|
Trav12n-3
|
T cell receptor alpha variable 12N-3 |
| chr2_-_88818544 | 0.00 |
ENSMUST00000099804.2
|
Olfr1214
|
olfactory receptor 1214 |
| chr2_-_89583170 | 0.00 |
ENSMUST00000099765.2
|
Olfr1253
|
olfactory receptor 1253 |
| chr10_+_77650454 | 0.00 |
ENSMUST00000220062.2
ENSMUST00000220393.2 |
Gm49918
|
predicted gene, 49918 |
| chr8_+_36956345 | 0.00 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr4_+_80752360 | 0.00 |
ENSMUST00000133655.8
ENSMUST00000006151.13 |
Tyrp1
|
tyrosinase-related protein 1 |
| chr19_-_39451509 | 0.00 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr17_-_40553176 | 0.00 |
ENSMUST00000026499.6
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr7_-_21863460 | 0.00 |
ENSMUST00000169697.2
|
Vmn1r142
|
vomeronasal 1 receptor 142 |
| chr10_+_129473353 | 0.00 |
ENSMUST00000214182.2
ENSMUST00000217364.2 ENSMUST00000216446.2 ENSMUST00000213820.2 |
Olfr799
|
olfactory receptor 799 |
| chr6_-_90050000 | 0.00 |
ENSMUST00000071865.2
|
Vmn1r49
|
vomeronasal 1, receptor 49 |
| chr18_-_60781365 | 0.00 |
ENSMUST00000143275.3
|
Synpo
|
synaptopodin |
| chr2_+_87820457 | 0.00 |
ENSMUST00000102622.2
|
Olfr1158
|
olfactory receptor 1158 |
| chr19_+_13448336 | 0.00 |
ENSMUST00000096202.2
|
Olfr1474
|
olfactory receptor 1474 |
| chr7_+_107916103 | 0.00 |
ENSMUST00000053179.2
|
Olfr491
|
olfactory receptor 491 |
| chr7_+_107413787 | 0.00 |
ENSMUST00000084756.2
|
Olfr467
|
olfactory receptor 467 |
| chr12_-_115019136 | 0.00 |
ENSMUST00000103519.2
ENSMUST00000192724.2 |
Ighv1-49
|
immunoglobulin heavy variable 1-49 |
| chr2_-_87958616 | 0.00 |
ENSMUST00000217575.3
|
Olfr1166
|
olfactory receptor 1166 |
| chr2_-_86369257 | 0.00 |
ENSMUST00000099882.2
|
Olfr1079
|
olfactory receptor 1079 |
| chr18_-_44292952 | 0.00 |
ENSMUST00000181652.9
|
Gm10267
|
predicted gene 10267 |
| chr7_-_21053786 | 0.00 |
ENSMUST00000105199.2
|
Vmn1r127
|
vomeronasal 1 receptor 127 |
| chr6_+_89870159 | 0.00 |
ENSMUST00000089420.6
|
Vmn1r44
|
vomeronasal 1 receptor 44 |
| chr7_-_20578910 | 0.00 |
ENSMUST00000169374.2
|
Vmn1r115
|
vomeronasal 1 receptor 115 |
| chr7_-_20868053 | 0.00 |
ENSMUST00000164409.2
|
Vmn1r122
|
vomeronasal 1 receptor 122 |
| chr6_+_56933451 | 0.00 |
ENSMUST00000096612.4
|
Vmn1r4
|
vomeronasal 1 receptor 4 |
| chr7_-_21496153 | 0.00 |
ENSMUST00000105196.2
|
Vmn1r248
|
vomeronasal 1 receptor 248 |
| chr7_-_22719348 | 0.00 |
ENSMUST00000168926.2
|
Vmn1r259
|
vomeronasal 1 receptor 259 |
| chr3_-_146302343 | 0.00 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr16_-_59184315 | 0.00 |
ENSMUST00000213779.2
ENSMUST00000208875.3 |
Olfr209
|
olfactory receptor 209 |
| chr16_-_19226828 | 0.00 |
ENSMUST00000052516.5
ENSMUST00000206410.3 |
Olfr165
|
olfactory receptor 165 |
| chr7_+_104529916 | 0.00 |
ENSMUST00000098165.2
|
Olfr665
|
olfactory receptor 665 |
| chr2_-_3367606 | 0.00 |
ENSMUST00000115087.4
ENSMUST00000027955.11 |
Olah
|
oleoyl-ACP hydrolase |
| chr4_-_112632013 | 0.00 |
ENSMUST00000060327.4
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr2_+_154004526 | 0.00 |
ENSMUST00000028986.3
|
Bpifa5
|
BPI fold containing family A, member 5 |
| chr7_+_19102423 | 0.00 |
ENSMUST00000132655.2
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_-_58647137 | 0.00 |
ENSMUST00000215069.2
ENSMUST00000079955.6 |
Olfr175
|
olfactory receptor 175 |
| chr7_+_29883611 | 0.00 |
ENSMUST00000208441.2
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr5_-_99184894 | 0.00 |
ENSMUST00000031277.7
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr7_+_103098253 | 0.00 |
ENSMUST00000217250.2
ENSMUST00000214631.2 |
Olfr606
|
olfactory receptor 606 |
| chr12_+_108376801 | 0.00 |
ENSMUST00000054955.14
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr4_+_114020581 | 0.00 |
ENSMUST00000079915.10
ENSMUST00000164297.8 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chr4_+_43851565 | 0.00 |
ENSMUST00000107860.3
|
Olfr155
|
olfactory receptor 155 |
| chr4_-_112632057 | 0.00 |
ENSMUST00000068851.6
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr2_+_36311412 | 0.00 |
ENSMUST00000071437.2
|
Olfr339
|
olfactory receptor 339 |
| chr3_-_129616136 | 0.00 |
ENSMUST00000029648.11
ENSMUST00000171313.8 ENSMUST00000200079.5 |
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
| chr18_+_37106851 | 0.00 |
ENSMUST00000192631.2
|
Pcdha7
|
protocadherin alpha 7 |
| chr14_+_65842716 | 0.00 |
ENSMUST00000079469.7
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr1_-_72739691 | 0.00 |
ENSMUST00000211837.2
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr4_+_48663504 | 0.00 |
ENSMUST00000030033.5
|
Cavin4
|
caveolae associated 4 |
| chr17_+_23819861 | 0.00 |
ENSMUST00000123866.8
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr13_-_22375311 | 0.00 |
ENSMUST00000238109.2
|
Vmn1r192
|
vomeronasal 1 receptor 192 |
| chr6_-_90201420 | 0.00 |
ENSMUST00000076086.3
|
Vmn1r53
|
vomeronasal 1 receptor 53 |
| chr14_+_53167246 | 0.00 |
ENSMUST00000177703.3
|
Trav12d-3
|
T cell receptor alpha variable 12D-3 |
| chr5_+_94224244 | 0.00 |
ENSMUST00000139102.2
|
Gm6351
|
predicted gene 6351 |
| chr8_-_121835125 | 0.00 |
ENSMUST00000133037.8
|
Mthfsd
|
methenyltetrahydrofolate synthetase domain containing |
| chr5_-_86345729 | 0.00 |
ENSMUST00000031172.9
ENSMUST00000113372.2 |
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr5_+_11552652 | 0.00 |
ENSMUST00000164651.8
|
Gm8906
|
predicted gene 8906 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0002582 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.0 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.0 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042282 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.0 | GO:0047316 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |