|
chr13_+_23715220
Show fit
|
0.61 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8
|
|
chr16_-_4698148
Show fit
|
0.54 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1
|
|
chr7_-_102607982
Show fit
|
0.53 |
ENSMUST00000216420.2
ENSMUST00000213477.2
|
Olfr575
|
olfactory receptor 575
|
|
chr8_-_33374282
Show fit
|
0.53 |
ENSMUST00000209107.4
ENSMUST00000209022.3
|
Nrg1
|
neuregulin 1
|
|
chr7_+_43000765
Show fit
|
0.53 |
ENSMUST00000012798.14
ENSMUST00000122423.8
|
Siglecf
|
sialic acid binding Ig-like lectin F
|
|
chr7_+_43000838
Show fit
|
0.42 |
ENSMUST00000206299.2
ENSMUST00000121494.2
|
Siglecf
|
sialic acid binding Ig-like lectin F
|
|
chr8_-_33374825
Show fit
|
0.41 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1
|
|
chr3_-_52012462
Show fit
|
0.40 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3
|
|
chr19_-_43974990
Show fit
|
0.40 |
ENSMUST00000026210.5
|
Cpn1
|
carboxypeptidase N, polypeptide 1
|
|
chr2_+_84810802
Show fit
|
0.36 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow
|
|
chr9_-_48747232
Show fit
|
0.35 |
ENSMUST00000093852.5
|
Zbtb16
|
zinc finger and BTB domain containing 16
|
|
chr2_-_168443540
Show fit
|
0.34 |
ENSMUST00000171689.8
ENSMUST00000137451.2
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2
|
|
chr2_+_19662529
Show fit
|
0.34 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1
|
|
chr13_-_21967540
Show fit
|
0.33 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11
|
|
chr7_+_101619897
Show fit
|
0.33 |
ENSMUST00000211272.2
|
Numa1
|
nuclear mitotic apparatus protein 1
|
|
chr12_-_31549538
Show fit
|
0.32 |
ENSMUST00000064240.14
ENSMUST00000185739.8
ENSMUST00000188326.3
ENSMUST00000101499.10
ENSMUST00000085487.12
|
Cbll1
|
Casitas B-lineage lymphoma-like 1
|
|
chr10_+_4432488
Show fit
|
0.31 |
ENSMUST00000138112.8
|
Ccdc170
|
coiled-coil domain containing 170
|
|
chr5_+_16758538
Show fit
|
0.30 |
ENSMUST00000199581.5
|
Hgf
|
hepatocyte growth factor
|
|
chr6_+_122803624
Show fit
|
0.30 |
ENSMUST00000203075.2
|
Foxj2
|
forkhead box J2
|
|
chrM_+_3906
Show fit
|
0.29 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2
|
|
chrX_-_20787150
Show fit
|
0.28 |
ENSMUST00000081893.7
ENSMUST00000115345.8
|
Syn1
|
synapsin I
|
|
chr7_+_110371811
Show fit
|
0.27 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr3_-_129834788
Show fit
|
0.27 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae)
|
|
chr5_+_146321757
Show fit
|
0.27 |
ENSMUST00000016143.9
|
Wasf3
|
WASP family, member 3
|
|
chr15_+_3300249
Show fit
|
0.26 |
ENSMUST00000082424.12
ENSMUST00000159158.9
ENSMUST00000159216.10
ENSMUST00000160311.3
|
Selenop
|
selenoprotein P
|
|
chr3_-_36107661
Show fit
|
0.26 |
ENSMUST00000166644.8
ENSMUST00000200469.5
|
Ccdc144b
|
coiled-coil domain containing 144B
|
|
chr8_+_66838927
Show fit
|
0.26 |
ENSMUST00000039540.12
ENSMUST00000110253.3
|
Marchf1
|
membrane associated ring-CH-type finger 1
|
|
chr13_-_98951627
Show fit
|
0.25 |
ENSMUST00000224992.2
ENSMUST00000225840.2
|
Fcho2
|
FCH domain only 2
|
|
chr5_-_66309244
Show fit
|
0.25 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47
|
|
chr6_-_135145129
Show fit
|
0.25 |
ENSMUST00000045855.9
|
Hebp1
|
heme binding protein 1
|
|
chr7_-_46569617
Show fit
|
0.25 |
ENSMUST00000210664.2
ENSMUST00000156335.9
|
Tsg101
|
tumor susceptibility gene 101
|
|
chr13_-_98951890
Show fit
|
0.25 |
ENSMUST00000040340.16
ENSMUST00000179563.8
ENSMUST00000109403.2
|
Fcho2
|
FCH domain only 2
|
|
chr6_+_79794899
Show fit
|
0.25 |
ENSMUST00000179797.3
|
Gm20594
|
predicted gene, 20594
|
|
chr5_+_16758777
Show fit
|
0.24 |
ENSMUST00000030683.8
|
Hgf
|
hepatocyte growth factor
|
|
chr1_-_69724939
Show fit
|
0.24 |
ENSMUST00000027146.9
|
Ikzf2
|
IKAROS family zinc finger 2
|
|
chr11_+_85243970
Show fit
|
0.24 |
ENSMUST00000108056.8
ENSMUST00000108061.8
ENSMUST00000108062.8
ENSMUST00000138423.8
ENSMUST00000092821.10
ENSMUST00000074875.11
|
Bcas3
|
breast carcinoma amplified sequence 3
|
|
chr11_-_115977755
Show fit
|
0.24 |
ENSMUST00000074628.13
ENSMUST00000106444.4
|
Wbp2
|
WW domain binding protein 2
|
|
chr3_-_95903313
Show fit
|
0.24 |
ENSMUST00000015889.10
|
Plekho1
|
pleckstrin homology domain containing, family O member 1
|
|
chr10_+_111342147
Show fit
|
0.23 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1
|
|
chr8_+_66070661
Show fit
|
0.23 |
ENSMUST00000110258.8
ENSMUST00000110256.8
ENSMUST00000110255.8
|
Marchf1
|
membrane associated ring-CH-type finger 1
|
|
chr5_+_16758897
Show fit
|
0.23 |
ENSMUST00000196645.2
|
Hgf
|
hepatocyte growth factor
|
|
chr16_-_36188086
Show fit
|
0.23 |
ENSMUST00000096089.3
|
Cstdc5
|
cystatin domain containing 5
|
|
chr7_-_80052491
Show fit
|
0.23 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr16_-_18884431
Show fit
|
0.23 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3
|
|
chr19_+_53317844
Show fit
|
0.22 |
ENSMUST00000111737.3
ENSMUST00000025998.15
ENSMUST00000237837.2
|
Mxi1
|
MAX interactor 1, dimerization protein
|
|
chr10_-_67972401
Show fit
|
0.22 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr8_-_64659004
Show fit
|
0.22 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like
|
|
chr6_-_99005212
Show fit
|
0.22 |
ENSMUST00000177437.8
ENSMUST00000177229.8
ENSMUST00000124058.8
|
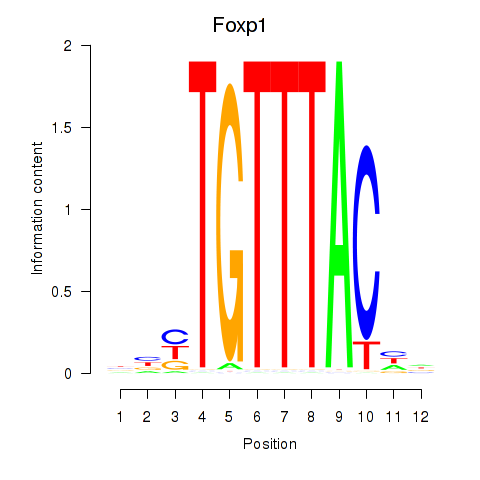
Foxp1
|
forkhead box P1
|
|
chr6_+_135339543
Show fit
|
0.22 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1
|
|
chr19_-_30526916
Show fit
|
0.22 |
ENSMUST00000025803.9
|
Dkk1
|
dickkopf WNT signaling pathway inhibitor 1
|
|
chr11_-_102446947
Show fit
|
0.22 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8
|
|
chr5_-_113968483
Show fit
|
0.21 |
ENSMUST00000100874.6
|
Selplg
|
selectin, platelet (p-selectin) ligand
|
|
chr17_-_34846323
Show fit
|
0.21 |
ENSMUST00000168709.3
ENSMUST00000064953.15
ENSMUST00000170345.8
ENSMUST00000171121.9
ENSMUST00000168391.9
ENSMUST00000169067.9
|
Gm20460
Ppt2
|
predicted gene 20460
palmitoyl-protein thioesterase 2
|
|
chr1_-_171023798
Show fit
|
0.21 |
ENSMUST00000111332.2
|
Pcp4l1
|
Purkinje cell protein 4-like 1
|
|
chr6_+_135339929
Show fit
|
0.21 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1
|
|
chr11_-_100288566
Show fit
|
0.20 |
ENSMUST00000001592.15
ENSMUST00000107403.2
|
Jup
|
junction plakoglobin
|
|
chr11_-_51647204
Show fit
|
0.20 |
ENSMUST00000109092.8
ENSMUST00000064297.5
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae)
|
|
chr2_+_152468850
Show fit
|
0.20 |
ENSMUST00000000369.4
ENSMUST00000150913.2
|
Rem1
|
rad and gem related GTP binding protein 1
|
|
chr6_-_39702127
Show fit
|
0.20 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene
|
|
chr9_-_39872429
Show fit
|
0.19 |
ENSMUST00000213171.2
|
Olfr976
|
olfactory receptor 976
|
|
chr18_-_80751327
Show fit
|
0.19 |
ENSMUST00000236310.2
ENSMUST00000167977.8
ENSMUST00000035800.8
|
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1
|
|
chr18_+_82928782
Show fit
|
0.19 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516
|
|
chr17_-_34846122
Show fit
|
0.19 |
ENSMUST00000171376.8
ENSMUST00000169287.2
|
Ppt2
|
palmitoyl-protein thioesterase 2
|
|
chr2_-_73722874
Show fit
|
0.19 |
ENSMUST00000136958.8
ENSMUST00000112010.9
ENSMUST00000128531.8
ENSMUST00000112017.8
|
Atf2
|
activating transcription factor 2
|
|
chr6_-_136781406
Show fit
|
0.19 |
ENSMUST00000179285.3
|
H4f16
|
H4 histone 16
|
|
chr8_-_122671588
Show fit
|
0.19 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial
|
|
chr11_+_116324913
Show fit
|
0.19 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2
|
|
chr15_+_102367463
Show fit
|
0.19 |
ENSMUST00000164938.8
ENSMUST00000023810.12
ENSMUST00000164957.8
ENSMUST00000171245.8
|
Prr13
|
proline rich 13
|
|
chr11_-_54846873
Show fit
|
0.19 |
ENSMUST00000155316.2
ENSMUST00000108889.10
ENSMUST00000126703.8
|
Tnip1
|
TNFAIP3 interacting protein 1
|
|
chr5_-_142803135
Show fit
|
0.19 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18
|
|
chr7_+_100435548
Show fit
|
0.18 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr8_+_71917554
Show fit
|
0.18 |
ENSMUST00000048914.8
|
Mrpl34
|
mitochondrial ribosomal protein L34
|
|
chr12_+_77284822
Show fit
|
0.18 |
ENSMUST00000177595.9
ENSMUST00000171770.10
|
Fut8
|
fucosyltransferase 8
|
|
chr19_+_8975249
Show fit
|
0.18 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin)
|
|
chr9_-_58648826
Show fit
|
0.18 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein
|
|
chr2_-_34261121
Show fit
|
0.18 |
ENSMUST00000127353.3
ENSMUST00000141653.3
|
Pbx3
|
pre B cell leukemia homeobox 3
|
|
chr3_-_137687284
Show fit
|
0.18 |
ENSMUST00000136613.4
ENSMUST00000029806.13
|
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1
|
|
chr13_-_104056803
Show fit
|
0.17 |
ENSMUST00000091269.11
ENSMUST00000188997.7
ENSMUST00000169083.8
ENSMUST00000191275.7
|
Erbin
|
Erbb2 interacting protein
|
|
chr16_-_59138611
Show fit
|
0.17 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204
|
|
chr2_+_69553141
Show fit
|
0.17 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G)
|
|
chr2_+_69553384
Show fit
|
0.17 |
ENSMUST00000040915.15
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G)
|
|
chr7_-_27252543
Show fit
|
0.17 |
ENSMUST00000127240.8
ENSMUST00000117095.8
ENSMUST00000117611.8
|
Pld3
|
phospholipase D family, member 3
|
|
chr8_+_73072877
Show fit
|
0.17 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung)
|
|
chr5_+_140491305
Show fit
|
0.17 |
ENSMUST00000043050.9
ENSMUST00000124142.2
|
Chst12
|
carbohydrate sulfotransferase 12
|
|
chr16_-_4376471
Show fit
|
0.17 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4
|
|
chr7_-_46569662
Show fit
|
0.17 |
ENSMUST00000143413.3
ENSMUST00000014546.15
|
Tsg101
|
tumor susceptibility gene 101
|
|
chr7_-_25176959
Show fit
|
0.16 |
ENSMUST00000098668.3
ENSMUST00000206687.2
ENSMUST00000206676.2
ENSMUST00000205308.2
ENSMUST00000098669.8
ENSMUST00000206171.2
ENSMUST00000098666.9
|
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1
|
|
chr8_-_70975734
Show fit
|
0.16 |
ENSMUST00000137610.3
|
Kxd1
|
KxDL motif containing 1
|
|
chr11_-_86884507
Show fit
|
0.16 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2
|
|
chr2_+_88660081
Show fit
|
0.16 |
ENSMUST00000215929.2
|
Olfr1205
|
olfactory receptor 1205
|
|
chr6_+_116627567
Show fit
|
0.16 |
ENSMUST00000067354.10
ENSMUST00000178241.4
|
Depp1
|
DEPP1 autophagy regulator
|
|
chr5_-_114127800
Show fit
|
0.16 |
ENSMUST00000077689.14
|
Ssh1
|
slingshot protein phosphatase 1
|
|
chr3_-_15397325
Show fit
|
0.16 |
ENSMUST00000108361.2
|
Gm9733
|
predicted gene 9733
|
|
chrX_+_93768175
Show fit
|
0.16 |
ENSMUST00000101388.4
|
Zxdb
|
zinc finger, X-linked, duplicated B
|
|
chr3_+_53396120
Show fit
|
0.16 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3
|
|
chr9_-_114811807
Show fit
|
0.15 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene
|
|
chr11_+_34264757
Show fit
|
0.15 |
ENSMUST00000165963.9
ENSMUST00000093192.4
|
Insyn2b
|
inhibitory synaptic factor family member 2B
|
|
chr1_+_173983199
Show fit
|
0.15 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420
|
|
chr17_+_46991972
Show fit
|
0.15 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1
|
|
chr8_-_70975813
Show fit
|
0.15 |
ENSMUST00000121623.8
ENSMUST00000093456.12
ENSMUST00000118850.8
|
Kxd1
|
KxDL motif containing 1
|
|
chr2_-_160714904
Show fit
|
0.15 |
ENSMUST00000109460.8
ENSMUST00000127201.2
|
Zhx3
|
zinc fingers and homeoboxes 3
|
|
chr6_-_57991216
Show fit
|
0.15 |
ENSMUST00000228951.2
|
Vmn1r26
|
vomeronasal 1 receptor 26
|
|
chr5_+_64969679
Show fit
|
0.15 |
ENSMUST00000166409.6
ENSMUST00000197879.2
|
Klf3
|
Kruppel-like factor 3 (basic)
|
|
chr11_+_118913788
Show fit
|
0.14 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2
|
|
chr5_+_122781941
Show fit
|
0.14 |
ENSMUST00000100737.10
ENSMUST00000121489.8
ENSMUST00000031425.15
ENSMUST00000086247.6
|
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr11_-_70129784
Show fit
|
0.14 |
ENSMUST00000141880.2
|
Rnasek
|
ribonuclease, RNase K
|
|
chr16_+_3408906
Show fit
|
0.14 |
ENSMUST00000216259.2
|
Olfr161
|
olfactory receptor 161
|
|
chr7_-_107633196
Show fit
|
0.14 |
ENSMUST00000210173.3
|
Olfr478
|
olfactory receptor 478
|
|
chr12_+_112645237
Show fit
|
0.14 |
ENSMUST00000174780.2
ENSMUST00000169593.2
ENSMUST00000173942.2
|
Zbtb42
|
zinc finger and BTB domain containing 42
|
|
chr6_+_89993129
Show fit
|
0.13 |
ENSMUST00000227811.2
ENSMUST00000228680.2
ENSMUST00000227229.2
|
Vmn1r47
|
vomeronasal 1 receptor 47
|
|
chr11_-_86698484
Show fit
|
0.13 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr4_-_55532453
Show fit
|
0.13 |
ENSMUST00000132746.2
ENSMUST00000107619.3
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr15_+_97682210
Show fit
|
0.13 |
ENSMUST00000117892.2
ENSMUST00000229084.2
|
Slc48a1
|
solute carrier family 48 (heme transporter), member 1
|
|
chr16_-_48592372
Show fit
|
0.13 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr6_-_99005835
Show fit
|
0.13 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1
|
|
chr10_+_60185093
Show fit
|
0.13 |
ENSMUST00000105459.2
|
Vsir
|
V-set immunoregulatory receptor
|
|
chr7_-_104570689
Show fit
|
0.13 |
ENSMUST00000217177.2
|
Olfr667
|
olfactory receptor 667
|
|
chr4_+_40269563
Show fit
|
0.13 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27
|
|
chr6_-_88851027
Show fit
|
0.13 |
ENSMUST00000038409.12
|
Podxl2
|
podocalyxin-like 2
|
|
chr4_-_128699838
Show fit
|
0.13 |
ENSMUST00000106072.9
ENSMUST00000170934.3
|
Zfp362
|
zinc finger protein 362
|
|
chr7_-_14297431
Show fit
|
0.13 |
ENSMUST00000227855.2
ENSMUST00000185220.3
ENSMUST00000226802.2
ENSMUST00000227692.2
ENSMUST00000227788.2
ENSMUST00000227566.2
ENSMUST00000226510.2
ENSMUST00000226264.2
|
Vmn1r90
|
vomeronasal 1 receptor 90
|
|
chr1_-_179373826
Show fit
|
0.12 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial
|
|
chr5_-_31065036
Show fit
|
0.12 |
ENSMUST00000132034.5
ENSMUST00000132253.5
|
Ost4
|
oligosaccharyltransferase complex subunit 4 (non-catalytic)
|
|
chr4_-_117354249
Show fit
|
0.12 |
ENSMUST00000030439.15
|
Rnf220
|
ring finger protein 220
|
|
chr15_-_66432938
Show fit
|
0.12 |
ENSMUST00000048372.7
|
Tmem71
|
transmembrane protein 71
|
|
chr14_+_60940394
Show fit
|
0.12 |
ENSMUST00000162131.9
|
Spata13
|
spermatogenesis associated 13
|
|
chr12_-_91745903
Show fit
|
0.12 |
ENSMUST00000170077.2
|
Ston2
|
stonin 2
|
|
chr11_-_70130620
Show fit
|
0.12 |
ENSMUST00000040428.4
|
Rnasek
|
ribonuclease, RNase K
|
|
chr7_-_106605189
Show fit
|
0.12 |
ENSMUST00000216375.2
ENSMUST00000208147.3
|
Olfr2
|
olfactory receptor 2
|
|
chr1_+_93918231
Show fit
|
0.12 |
ENSMUST00000097632.5
|
Gal3st2c
|
galactose-3-O-sulfotransferase 2C
|
|
chr6_-_5496261
Show fit
|
0.12 |
ENSMUST00000203347.3
ENSMUST00000019721.7
|
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4
|
|
chr1_+_172525613
Show fit
|
0.12 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related
|
|
chr10_+_93324624
Show fit
|
0.12 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase
|
|
chr4_-_154245073
Show fit
|
0.12 |
ENSMUST00000105639.4
ENSMUST00000030896.15
|
Tprgl
|
transformation related protein 63 regulated like
|
|
chr6_-_99073156
Show fit
|
0.12 |
ENSMUST00000175886.8
|
Foxp1
|
forkhead box P1
|
|
chr2_+_112096154
Show fit
|
0.12 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr2_+_87610895
Show fit
|
0.12 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152
|
|
chr5_+_31046196
Show fit
|
0.12 |
ENSMUST00000069705.11
ENSMUST00000201917.4
ENSMUST00000201168.4
ENSMUST00000202060.4
ENSMUST00000201225.4
ENSMUST00000114700.9
|
Agbl5
|
ATP/GTP binding protein-like 5
|
|
chr7_-_44498305
Show fit
|
0.12 |
ENSMUST00000207293.2
ENSMUST00000207532.2
|
Tbc1d17
|
TBC1 domain family, member 17
|
|
chr15_+_79232137
Show fit
|
0.12 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian)
|
|
chr1_-_44258112
Show fit
|
0.12 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E
|
|
chr15_+_102368510
Show fit
|
0.12 |
ENSMUST00000164688.2
|
Prr13
|
proline rich 13
|
|
chr4_-_15149755
Show fit
|
0.12 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1
|
|
chr10_-_57408585
Show fit
|
0.12 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1
|
|
chr17_-_37938000
Show fit
|
0.11 |
ENSMUST00000223366.2
ENSMUST00000216128.2
|
Olfr115
Olfr116
|
olfactory receptor 115
olfactory receptor 116
|
|
chr14_+_54664359
Show fit
|
0.11 |
ENSMUST00000010550.12
ENSMUST00000199195.3
ENSMUST00000196273.2
|
Mrpl52
|
mitochondrial ribosomal protein L52
|
|
chr7_+_140181182
Show fit
|
0.11 |
ENSMUST00000214180.2
ENSMUST00000211771.2
|
Olfr46
|
olfactory receptor 46
|
|
chr17_+_46991906
Show fit
|
0.11 |
ENSMUST00000233430.2
|
Mea1
|
male enhanced antigen 1
|
|
chr16_+_48692976
Show fit
|
0.11 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma
|
|
chr17_-_38618461
Show fit
|
0.11 |
ENSMUST00000213505.2
|
Olfr137
|
olfactory receptor 137
|
|
chr17_+_56920389
Show fit
|
0.11 |
ENSMUST00000080492.7
|
Rpl36
|
ribosomal protein L36
|
|
chr7_-_29204812
Show fit
|
0.11 |
ENSMUST00000183096.8
ENSMUST00000085809.11
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3
|
|
chr14_-_122153185
Show fit
|
0.11 |
ENSMUST00000055475.9
|
Gpr18
|
G protein-coupled receptor 18
|
|
chrX_+_158623460
Show fit
|
0.11 |
ENSMUST00000112451.8
ENSMUST00000112453.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_134343152
Show fit
|
0.11 |
ENSMUST00000112237.2
|
Adipor1
|
adiponectin receptor 1
|
|
chr17_+_47815998
Show fit
|
0.11 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3
|
|
chr10_+_80084955
Show fit
|
0.11 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7
|
|
chr19_+_23118545
Show fit
|
0.11 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9
|
|
chr6_-_88851579
Show fit
|
0.11 |
ENSMUST00000061262.11
ENSMUST00000140455.8
ENSMUST00000145780.2
|
Podxl2
|
podocalyxin-like 2
|
|
chr1_+_34044940
Show fit
|
0.11 |
ENSMUST00000187486.7
ENSMUST00000182697.8
|
Dst
|
dystonin
|
|
chr8_+_65399831
Show fit
|
0.11 |
ENSMUST00000026595.13
ENSMUST00000209852.2
ENSMUST00000079896.9
|
Tmem192
|
transmembrane protein 192
|
|
chr2_+_127967951
Show fit
|
0.11 |
ENSMUST00000089634.12
ENSMUST00000019281.14
ENSMUST00000110341.9
ENSMUST00000103211.8
ENSMUST00000103210.2
|
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr10_+_127256736
Show fit
|
0.11 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2
|
|
chr11_-_106606076
Show fit
|
0.11 |
ENSMUST00000080853.11
ENSMUST00000183610.8
ENSMUST00000103069.10
ENSMUST00000106796.9
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr1_+_107456731
Show fit
|
0.11 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr5_-_124003553
Show fit
|
0.11 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2
|
|
chr8_+_120729459
Show fit
|
0.11 |
ENSMUST00000108972.4
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr7_+_46700349
Show fit
|
0.11 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A
|
|
chr15_-_58078274
Show fit
|
0.10 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32
|
|
chr9_+_38119661
Show fit
|
0.10 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893
|
|
chr6_+_58278534
Show fit
|
0.10 |
ENSMUST00000228909.2
ENSMUST00000227761.2
ENSMUST00000228038.2
ENSMUST00000226971.2
|
Vmn1r29
|
vomeronasal 1 receptor 29
|
|
chr3_+_90161470
Show fit
|
0.10 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2
|
|
chr4_-_82803384
Show fit
|
0.10 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist
|
|
chr12_-_32000169
Show fit
|
0.10 |
ENSMUST00000176520.8
|
Hbp1
|
high mobility group box transcription factor 1
|
|
chr9_-_62895197
Show fit
|
0.10 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1
|
|
chr1_+_134343107
Show fit
|
0.10 |
ENSMUST00000027727.15
|
Adipor1
|
adiponectin receptor 1
|
|
chr1_-_92569663
Show fit
|
0.10 |
ENSMUST00000097642.4
|
Cops9
|
COP9 signalosome subunit 9
|
|
chr2_+_31985528
Show fit
|
0.10 |
ENSMUST00000057423.6
ENSMUST00000140762.2
|
Plpp7
|
phospholipid phosphatase 7 (inactive)
|
|
chr1_+_88030951
Show fit
|
0.10 |
ENSMUST00000113135.6
ENSMUST00000113138.8
|
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr5_-_136595205
Show fit
|
0.10 |
ENSMUST00000176778.8
|
Cux1
|
cut-like homeobox 1
|
|
chr7_+_3339059
Show fit
|
0.10 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr15_-_50746202
Show fit
|
0.10 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1
|
|
chr19_-_34856853
Show fit
|
0.10 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1
|
|
chr1_-_139304779
Show fit
|
0.10 |
ENSMUST00000196402.5
ENSMUST00000059825.12
|
Crb1
|
crumbs family member 1, photoreceptor morphogenesis associated
|
|
chr2_-_59955995
Show fit
|
0.10 |
ENSMUST00000112550.8
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr9_-_58156982
Show fit
|
0.10 |
ENSMUST00000135310.8
ENSMUST00000085673.11
ENSMUST00000114136.9
ENSMUST00000153820.8
ENSMUST00000124982.2
|
Pml
|
promyelocytic leukemia
|
|
chr2_-_104324035
Show fit
|
0.10 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3
|
|
chr9_-_49479791
Show fit
|
0.10 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr15_+_79231720
Show fit
|
0.10 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian)
|
|
chr11_+_54329014
Show fit
|
0.10 |
ENSMUST00000046835.14
|
Fnip1
|
folliculin interacting protein 1
|
|
chr3_+_146205864
Show fit
|
0.10 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5
|
|
chr10_-_5144699
Show fit
|
0.10 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr18_-_39622932
Show fit
|
0.10 |
ENSMUST00000152853.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1
|
|
chr8_+_95721378
Show fit
|
0.10 |
ENSMUST00000212956.2
ENSMUST00000212531.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1
|
|
chr9_+_50405817
Show fit
|
0.09 |
ENSMUST00000114474.8
ENSMUST00000188047.2
|
Plet1
|
placenta expressed transcript 1
|
|
chr7_+_3339077
Show fit
|
0.09 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr16_-_23807602
Show fit
|
0.09 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6
|
|
chr15_+_61857226
Show fit
|
0.09 |
ENSMUST00000161976.8
ENSMUST00000022971.8
|
Myc
|
myelocytomatosis oncogene
|
|
chr17_+_43978377
Show fit
|
0.09 |
ENSMUST00000233627.2
ENSMUST00000233437.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1
|
|
chr9_-_58156935
Show fit
|
0.09 |
ENSMUST00000124063.2
ENSMUST00000126690.8
|
Pml
|
promyelocytic leukemia
|