Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
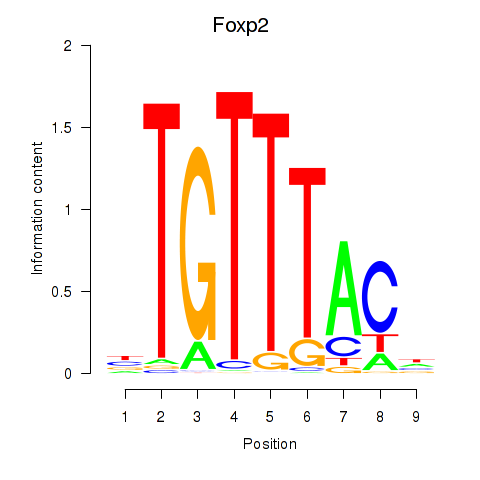
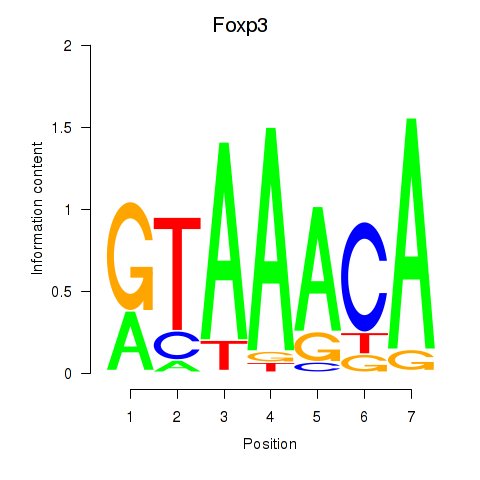
Results for Foxp2_Foxp3
Z-value: 0.92
Transcription factors associated with Foxp2_Foxp3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxp2
|
ENSMUSG00000029563.17 | forkhead box P2 |
|
Foxp3
|
ENSMUSG00000039521.14 | forkhead box P3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxp2 | mm39_v1_chr6_+_14901343_14901363 | 0.96 | 1.1e-02 | Click! |
| Foxp3 | mm39_v1_chrX_+_7439839_7439883 | 0.89 | 4.3e-02 | Click! |
Activity profile of Foxp2_Foxp3 motif
Sorted Z-values of Foxp2_Foxp3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_79794899 | 1.74 |
ENSMUST00000179797.3
|
Gm20594
|
predicted gene, 20594 |
| chr2_-_168443540 | 1.19 |
ENSMUST00000171689.8
ENSMUST00000137451.2 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr16_-_4698148 | 1.13 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr6_+_116627567 | 1.11 |
ENSMUST00000067354.10
ENSMUST00000178241.4 |
Depp1
|
DEPP1 autophagy regulator |
| chr13_-_23946359 | 1.01 |
ENSMUST00000091701.3
|
H3c1
|
H3 clustered histone 1 |
| chr11_+_68986043 | 0.90 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr13_-_98951627 | 0.88 |
ENSMUST00000224992.2
ENSMUST00000225840.2 |
Fcho2
|
FCH domain only 2 |
| chr8_-_64659004 | 0.82 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr3_-_52012462 | 0.81 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3 |
| chr3_-_129834788 | 0.80 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr1_+_34044940 | 0.73 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr18_-_80751327 | 0.70 |
ENSMUST00000236310.2
ENSMUST00000167977.8 ENSMUST00000035800.8 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr13_-_98951890 | 0.66 |
ENSMUST00000040340.16
ENSMUST00000179563.8 ENSMUST00000109403.2 |
Fcho2
|
FCH domain only 2 |
| chr13_-_21967540 | 0.65 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr5_-_142803135 | 0.65 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr7_+_110371811 | 0.63 |
ENSMUST00000005829.13
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr13_+_23930717 | 0.63 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3 |
| chr1_-_64995982 | 0.62 |
ENSMUST00000097713.2
|
Plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr16_-_4376471 | 0.62 |
ENSMUST00000230875.2
|
Tfap4
|
transcription factor AP4 |
| chr8_+_66838927 | 0.61 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr15_-_5137975 | 0.60 |
ENSMUST00000118365.3
|
Card6
|
caspase recruitment domain family, member 6 |
| chr10_-_67972401 | 0.60 |
ENSMUST00000218532.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr12_-_31549538 | 0.57 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr8_-_122671588 | 0.55 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr11_-_86884507 | 0.55 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr3_-_102871440 | 0.54 |
ENSMUST00000058899.13
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr14_+_61844899 | 0.50 |
ENSMUST00000225582.2
ENSMUST00000051184.10 |
Kcnrg
|
potassium channel regulator |
| chr10_+_29087602 | 0.50 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chrM_-_14061 | 0.50 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr11_+_85243970 | 0.49 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chr6_+_15185202 | 0.49 |
ENSMUST00000154448.2
|
Foxp2
|
forkhead box P2 |
| chr6_+_116627635 | 0.49 |
ENSMUST00000204555.2
|
Depp1
|
DEPP1 autophagy regulator |
| chr3_+_53396120 | 0.49 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr6_-_39702127 | 0.47 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr10_+_59942274 | 0.47 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr11_-_102255999 | 0.46 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr4_-_19922599 | 0.44 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr11_+_70104929 | 0.44 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr15_+_3300249 | 0.44 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr11_+_34264757 | 0.42 |
ENSMUST00000165963.9
ENSMUST00000093192.4 |
Insyn2b
|
inhibitory synaptic factor family member 2B |
| chr11_-_54846873 | 0.42 |
ENSMUST00000155316.2
ENSMUST00000108889.10 ENSMUST00000126703.8 |
Tnip1
|
TNFAIP3 interacting protein 1 |
| chr11_-_102446947 | 0.42 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8 |
| chr4_-_82803384 | 0.38 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr1_+_171052623 | 0.37 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr1_+_16758629 | 0.36 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr10_-_130002635 | 0.35 |
ENSMUST00000216530.2
|
Olfr825
|
olfactory receptor 825 |
| chr8_-_33374282 | 0.35 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chrX_-_133062677 | 0.35 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr19_-_27988393 | 0.34 |
ENSMUST00000172907.8
ENSMUST00000046898.17 |
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr3_+_90161470 | 0.34 |
ENSMUST00000029545.15
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr11_+_107438751 | 0.34 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr5_+_3978032 | 0.33 |
ENSMUST00000143365.8
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr10_+_93324624 | 0.33 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr14_+_48358267 | 0.33 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr19_-_12313274 | 0.33 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chrX_+_150303650 | 0.33 |
ENSMUST00000112666.8
ENSMUST00000112662.9 ENSMUST00000168501.8 |
Phf8
|
PHD finger protein 8 |
| chr17_+_47815998 | 0.32 |
ENSMUST00000183210.2
|
Ccnd3
|
cyclin D3 |
| chr17_+_75742881 | 0.32 |
ENSMUST00000164192.9
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr3_+_60380243 | 0.31 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr6_-_99005212 | 0.31 |
ENSMUST00000177437.8
ENSMUST00000177229.8 ENSMUST00000124058.8 |
Foxp1
|
forkhead box P1 |
| chr11_-_101117237 | 0.31 |
ENSMUST00000100417.3
ENSMUST00000107285.8 ENSMUST00000107284.8 |
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr12_-_32000534 | 0.30 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr2_+_145627900 | 0.30 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr5_+_64969679 | 0.30 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr6_-_5496261 | 0.30 |
ENSMUST00000203347.3
ENSMUST00000019721.7 |
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4 |
| chr2_-_73722874 | 0.30 |
ENSMUST00000136958.8
ENSMUST00000112010.9 ENSMUST00000128531.8 ENSMUST00000112017.8 |
Atf2
|
activating transcription factor 2 |
| chrX_+_102400061 | 0.30 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_-_128520002 | 0.30 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr1_+_173983199 | 0.30 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr19_-_27988534 | 0.30 |
ENSMUST00000174850.8
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr6_+_136931519 | 0.28 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr1_+_172525613 | 0.28 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr1_-_156767123 | 0.28 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_-_34846323 | 0.28 |
ENSMUST00000168709.3
ENSMUST00000064953.15 ENSMUST00000170345.8 ENSMUST00000171121.9 ENSMUST00000168391.9 ENSMUST00000169067.9 |
Gm20460
Ppt2
|
predicted gene 20460 palmitoyl-protein thioesterase 2 |
| chr6_-_99073156 | 0.28 |
ENSMUST00000175886.8
|
Foxp1
|
forkhead box P1 |
| chr3_-_133250889 | 0.27 |
ENSMUST00000197118.5
|
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr9_+_57818243 | 0.27 |
ENSMUST00000216925.2
ENSMUST00000163329.2 ENSMUST00000213654.2 ENSMUST00000217132.2 ENSMUST00000216841.2 ENSMUST00000214086.2 |
Ubl7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr11_-_100288566 | 0.27 |
ENSMUST00000001592.15
ENSMUST00000107403.2 |
Jup
|
junction plakoglobin |
| chr16_-_48592372 | 0.27 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr11_-_58521327 | 0.26 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr7_-_80052491 | 0.26 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr14_-_71004019 | 0.26 |
ENSMUST00000167242.8
|
Xpo7
|
exportin 7 |
| chr10_+_127512933 | 0.26 |
ENSMUST00000118612.8
ENSMUST00000048099.5 |
Nemp1
|
nuclear envelope integral membrane protein 1 |
| chr15_+_8997480 | 0.26 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr10_+_85763545 | 0.26 |
ENSMUST00000170396.3
|
Ascl4
|
achaete-scute family bHLH transcription factor 4 |
| chr2_+_112096154 | 0.26 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr2_-_172782089 | 0.26 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr17_-_34846122 | 0.26 |
ENSMUST00000171376.8
ENSMUST00000169287.2 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr8_-_33374825 | 0.26 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1 |
| chr5_+_139375540 | 0.25 |
ENSMUST00000100514.3
|
Gpr146
|
G protein-coupled receptor 146 |
| chr11_+_70104736 | 0.25 |
ENSMUST00000171032.8
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr19_+_4203603 | 0.25 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr8_+_66070661 | 0.25 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr15_-_80929669 | 0.24 |
ENSMUST00000135047.2
|
Mrtfa
|
myocardin related transcription factor A |
| chr11_-_74480870 | 0.24 |
ENSMUST00000145524.2
ENSMUST00000102521.9 |
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr2_+_19662529 | 0.24 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr19_+_13339600 | 0.24 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr5_+_104447037 | 0.23 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr19_+_5927876 | 0.23 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr9_+_119231140 | 0.23 |
ENSMUST00000165044.3
|
Acvr2b
|
activin receptor IIB |
| chr17_+_17669082 | 0.23 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr18_-_78166595 | 0.23 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr12_-_100691316 | 0.23 |
ENSMUST00000222731.2
|
Rps6ka5
|
ribosomal protein S6 kinase, polypeptide 5 |
| chr2_+_69553384 | 0.22 |
ENSMUST00000040915.15
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr2_+_136555364 | 0.22 |
ENSMUST00000028727.11
ENSMUST00000110098.4 |
Snap25
|
synaptosomal-associated protein 25 |
| chr3_-_36107661 | 0.22 |
ENSMUST00000166644.8
ENSMUST00000200469.5 |
Ccdc144b
|
coiled-coil domain containing 144B |
| chr9_-_37627519 | 0.22 |
ENSMUST00000215727.2
ENSMUST00000211952.3 |
Olfr160
|
olfactory receptor 160 |
| chr15_+_61857226 | 0.22 |
ENSMUST00000161976.8
ENSMUST00000022971.8 |
Myc
|
myelocytomatosis oncogene |
| chr11_+_110858842 | 0.22 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_-_66309244 | 0.22 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr4_-_128699838 | 0.22 |
ENSMUST00000106072.9
ENSMUST00000170934.3 |
Zfp362
|
zinc finger protein 362 |
| chr2_-_73722932 | 0.22 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr7_+_100435548 | 0.22 |
ENSMUST00000216021.2
|
Fam168a
|
family with sequence similarity 168, member A |
| chr7_-_44498305 | 0.22 |
ENSMUST00000207293.2
ENSMUST00000207532.2 |
Tbc1d17
|
TBC1 domain family, member 17 |
| chr9_-_39486831 | 0.22 |
ENSMUST00000216298.2
ENSMUST00000215194.2 |
Olfr959
|
olfactory receptor 959 |
| chr2_-_153079828 | 0.21 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr14_-_52481907 | 0.21 |
ENSMUST00000226307.2
|
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr19_-_43974990 | 0.21 |
ENSMUST00000026210.5
|
Cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr7_-_103674780 | 0.21 |
ENSMUST00000218535.2
|
Olfr640
|
olfactory receptor 640 |
| chr14_+_8283087 | 0.21 |
ENSMUST00000206298.3
ENSMUST00000216079.2 |
Olfr720
|
olfactory receptor 720 |
| chr7_+_101619897 | 0.21 |
ENSMUST00000211272.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr1_-_30912916 | 0.21 |
ENSMUST00000188780.2
|
Phf3
|
PHD finger protein 3 |
| chr3_+_137573839 | 0.21 |
ENSMUST00000197711.2
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr1_+_130659700 | 0.20 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr19_-_7318798 | 0.20 |
ENSMUST00000165965.8
ENSMUST00000051711.16 ENSMUST00000169541.8 ENSMUST00000165989.2 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr18_+_37453427 | 0.20 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr2_+_69553141 | 0.20 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G) |
| chr2_+_31864438 | 0.20 |
ENSMUST00000065398.13
|
Nup214
|
nucleoporin 214 |
| chr1_+_37338964 | 0.20 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr13_-_95661726 | 0.20 |
ENSMUST00000022185.10
|
F2rl1
|
coagulation factor II (thrombin) receptor-like 1 |
| chrM_+_11735 | 0.20 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr13_-_93636224 | 0.19 |
ENSMUST00000220513.2
ENSMUST00000065537.9 |
Jmy
|
junction-mediating and regulatory protein |
| chr4_-_123507494 | 0.19 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr7_+_140641010 | 0.19 |
ENSMUST00000048002.7
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr7_+_44499005 | 0.19 |
ENSMUST00000150335.2
ENSMUST00000107882.8 |
Akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr12_-_72117865 | 0.19 |
ENSMUST00000050649.6
|
Gpr135
|
G protein-coupled receptor 135 |
| chr6_-_106725929 | 0.19 |
ENSMUST00000204659.3
|
Il5ra
|
interleukin 5 receptor, alpha |
| chr2_-_36372480 | 0.19 |
ENSMUST00000213300.2
|
Olfr341
|
olfactory receptor 341 |
| chr7_-_29204812 | 0.19 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr10_+_63222338 | 0.19 |
ENSMUST00000043317.7
|
Dnajc12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chrX_+_158623460 | 0.19 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr4_+_100336003 | 0.19 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr12_+_119356318 | 0.19 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1 |
| chr12_+_71063431 | 0.18 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr13_-_53083674 | 0.18 |
ENSMUST00000120535.8
ENSMUST00000119311.8 ENSMUST00000021913.16 ENSMUST00000110031.4 |
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr12_-_32000209 | 0.18 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chr4_+_119590971 | 0.18 |
ENSMUST00000084306.5
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr11_+_116324913 | 0.18 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2 |
| chr8_+_121854566 | 0.18 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr17_+_46991972 | 0.18 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr2_-_73284262 | 0.18 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_139304779 | 0.18 |
ENSMUST00000196402.5
ENSMUST00000059825.12 |
Crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr15_-_50746202 | 0.17 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr11_+_54329014 | 0.17 |
ENSMUST00000046835.14
|
Fnip1
|
folliculin interacting protein 1 |
| chr2_-_172296662 | 0.17 |
ENSMUST00000161334.2
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr2_+_127967951 | 0.17 |
ENSMUST00000089634.12
ENSMUST00000019281.14 ENSMUST00000110341.9 ENSMUST00000103211.8 ENSMUST00000103210.2 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr9_-_58648826 | 0.17 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr18_-_39622932 | 0.17 |
ENSMUST00000152853.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr6_-_39534765 | 0.17 |
ENSMUST00000036877.10
ENSMUST00000154149.2 |
Dennd2a
|
DENN/MADD domain containing 2A |
| chr6_-_87312743 | 0.17 |
ENSMUST00000042025.12
ENSMUST00000205033.2 |
Antxr1
|
anthrax toxin receptor 1 |
| chr2_-_103858632 | 0.16 |
ENSMUST00000056170.4
|
4931422A03Rik
|
RIKEN cDNA 4931422A03 gene |
| chr10_+_4432488 | 0.16 |
ENSMUST00000138112.8
|
Ccdc170
|
coiled-coil domain containing 170 |
| chr1_-_54965470 | 0.16 |
ENSMUST00000179030.8
ENSMUST00000044359.16 |
Ankrd44
|
ankyrin repeat domain 44 |
| chr16_+_43330630 | 0.16 |
ENSMUST00000114695.3
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_+_107497109 | 0.16 |
ENSMUST00000209670.4
ENSMUST00000216937.3 |
Olfr472
|
olfactory receptor 472 |
| chr3_+_101917392 | 0.16 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr5_-_142803405 | 0.16 |
ENSMUST00000151477.8
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr11_+_96820091 | 0.16 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr19_-_34856853 | 0.16 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr11_+_97206542 | 0.16 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr17_+_29042640 | 0.16 |
ENSMUST00000233088.2
ENSMUST00000233182.2 ENSMUST00000233520.2 |
Brpf3
|
bromodomain and PHD finger containing, 3 |
| chr15_-_36609812 | 0.16 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_+_38312994 | 0.16 |
ENSMUST00000214648.3
ENSMUST00000056364.3 |
Olfr147
|
olfactory receptor 147 |
| chr1_-_169938060 | 0.16 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr10_-_66932685 | 0.15 |
ENSMUST00000020023.9
|
Reep3
|
receptor accessory protein 3 |
| chr6_-_41681273 | 0.15 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr19_-_7319157 | 0.15 |
ENSMUST00000164205.8
ENSMUST00000165286.8 ENSMUST00000168324.2 ENSMUST00000032557.15 |
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr3_+_84500854 | 0.15 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr13_+_49697919 | 0.15 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr19_-_47303184 | 0.15 |
ENSMUST00000237796.2
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr3_-_37286714 | 0.15 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr3_+_60380463 | 0.15 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr11_+_69657275 | 0.15 |
ENSMUST00000132528.8
ENSMUST00000153943.2 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr18_+_82928782 | 0.15 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr13_+_37529184 | 0.15 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr19_-_29721012 | 0.15 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr2_+_24235300 | 0.15 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr13_+_81034214 | 0.15 |
ENSMUST00000161441.2
|
Arrdc3
|
arrestin domain containing 3 |
| chr9_-_102231884 | 0.14 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr17_-_38618461 | 0.14 |
ENSMUST00000213505.2
|
Olfr137
|
olfactory receptor 137 |
| chr12_-_72283465 | 0.14 |
ENSMUST00000021497.16
ENSMUST00000137990.2 |
Rtn1
|
reticulon 1 |
| chr4_-_148584906 | 0.14 |
ENSMUST00000030840.4
|
Angptl7
|
angiopoietin-like 7 |
| chr10_-_95159933 | 0.14 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr18_+_37580692 | 0.14 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr13_+_42205790 | 0.14 |
ENSMUST00000220525.2
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr10_-_12424623 | 0.14 |
ENSMUST00000219003.2
|
Utrn
|
utrophin |
| chr8_-_32408380 | 0.14 |
ENSMUST00000208497.3
ENSMUST00000207584.3 |
Nrg1
|
neuregulin 1 |
| chr2_-_45000389 | 0.14 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_34251041 | 0.14 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta |
| chr2_+_113235475 | 0.14 |
ENSMUST00000238883.2
|
Fmn1
|
formin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxp2_Foxp3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.2 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.6 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.2 | 0.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.5 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.5 | GO:0061187 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.4 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.4 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.1 | 0.9 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.3 | GO:0046722 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.3 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.1 | 0.2 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.6 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.7 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.2 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.2 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.5 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0061033 | fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 1.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.8 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.1 | GO:0033624 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.0 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.6 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:2000771 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0072023 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.0 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:2001015 | skeletal muscle satellite cell activation(GO:0014719) negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0071655 | negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.0 | 0.2 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 0.1 | GO:2000656 | MAPK import into nucleus(GO:0000189) regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.0 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.1 | 0.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.7 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.1 | 0.4 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |