Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
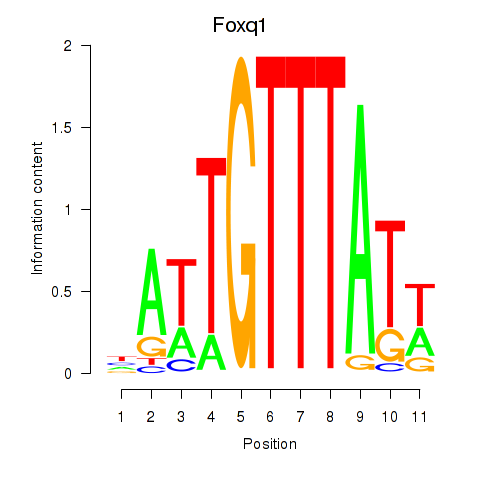
Results for Foxq1
Z-value: 0.39
Transcription factors associated with Foxq1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxq1
|
ENSMUSG00000038415.11 | forkhead box Q1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxq1 | mm39_v1_chr13_+_31740117_31740117 | -0.93 | 2.4e-02 | Click! |
Activity profile of Foxq1 motif
Sorted Z-values of Foxq1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_18910340 | 0.15 |
ENSMUST00000117338.8
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr18_+_13107535 | 0.15 |
ENSMUST00000234035.2
ENSMUST00000235053.2 |
Impact
|
impact, RWD domain protein |
| chr5_-_121025645 | 0.14 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr3_+_129326004 | 0.13 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr4_-_87724533 | 0.12 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr19_+_43770619 | 0.12 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr12_-_73160181 | 0.11 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chrX_+_138464065 | 0.11 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr15_-_55411560 | 0.10 |
ENSMUST00000165356.2
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr2_+_156681991 | 0.10 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr10_+_34173426 | 0.10 |
ENSMUST00000047935.8
|
Tspyl4
|
TSPY-like 4 |
| chr6_+_38895902 | 0.09 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr3_+_31150982 | 0.09 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr4_-_87724512 | 0.09 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr2_+_173579285 | 0.09 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr5_-_115236354 | 0.09 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr3_+_96552895 | 0.09 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chr17_+_30845909 | 0.09 |
ENSMUST00000236140.2
ENSMUST00000236118.2 ENSMUST00000235390.2 |
Dnah8
|
dynein, axonemal, heavy chain 8 |
| chr9_-_105398346 | 0.08 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr19_+_45433899 | 0.08 |
ENSMUST00000224478.2
|
Btrc
|
beta-transducin repeat containing protein |
| chr19_-_36896999 | 0.08 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr2_+_156681927 | 0.08 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr13_+_28441511 | 0.08 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chrX_+_37689503 | 0.08 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr9_-_100388857 | 0.08 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr18_+_69477541 | 0.08 |
ENSMUST00000114985.10
ENSMUST00000128706.8 ENSMUST00000201781.4 ENSMUST00000202674.4 |
Tcf4
|
transcription factor 4 |
| chr7_+_16807965 | 0.08 |
ENSMUST00000071399.13
ENSMUST00000118367.3 |
Psg16
|
pregnancy specific glycoprotein 16 |
| chr15_-_102097466 | 0.07 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr11_+_96920751 | 0.07 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr11_-_82719850 | 0.07 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr8_-_54091980 | 0.07 |
ENSMUST00000047768.11
|
Neil3
|
nei like 3 (E. coli) |
| chr5_-_92496730 | 0.07 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr18_+_11790409 | 0.07 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr4_-_15945359 | 0.07 |
ENSMUST00000029877.9
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr2_-_25498459 | 0.07 |
ENSMUST00000058137.9
|
Rabl6
|
RAB, member RAS oncogene family-like 6 |
| chr3_+_135531409 | 0.07 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr5_+_104447037 | 0.06 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr7_+_79836581 | 0.06 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr9_+_108437485 | 0.06 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr19_-_29721012 | 0.06 |
ENSMUST00000175764.9
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr8_+_79236051 | 0.06 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr11_-_107238956 | 0.06 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr1_+_43137852 | 0.06 |
ENSMUST00000010434.8
|
AI597479
|
expressed sequence AI597479 |
| chr6_+_123149847 | 0.06 |
ENSMUST00000088455.6
|
Clec4b2
|
C-type lectin domain family 4, member b2 |
| chr12_-_83643883 | 0.06 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr5_-_138169253 | 0.05 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr17_+_19724994 | 0.05 |
ENSMUST00000166081.3
ENSMUST00000231465.2 |
Vmn2r100
|
vomeronasal 2, receptor 100 |
| chr19_-_47452840 | 0.05 |
ENSMUST00000081619.10
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr1_+_87332638 | 0.05 |
ENSMUST00000173152.2
ENSMUST00000173663.2 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr16_-_92196954 | 0.05 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr19_-_43663282 | 0.05 |
ENSMUST00000046038.9
ENSMUST00000236433.2 |
Slc25a28
|
solute carrier family 25, member 28 |
| chr13_-_32967937 | 0.05 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr3_+_129326285 | 0.05 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chrX_-_156351979 | 0.05 |
ENSMUST00000065806.5
|
Yy2
|
Yy2 transcription factor |
| chr17_-_35265702 | 0.05 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr17_-_30845845 | 0.05 |
ENSMUST00000235547.2
ENSMUST00000188852.2 |
Glo1
1700097N02Rik
|
glyoxalase 1 RIKEN cDNA 1700097N02 gene |
| chr4_+_139350152 | 0.05 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr9_-_56151334 | 0.05 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr5_+_24679154 | 0.04 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr14_-_54651442 | 0.04 |
ENSMUST00000227334.2
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr1_-_45542442 | 0.04 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr4_+_132366298 | 0.04 |
ENSMUST00000135299.8
ENSMUST00000020197.14 ENSMUST00000180250.8 ENSMUST00000081726.13 ENSMUST00000079157.11 |
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr5_-_124717146 | 0.04 |
ENSMUST00000031334.15
|
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chr1_-_44140820 | 0.04 |
ENSMUST00000152239.8
|
Tex30
|
testis expressed 30 |
| chr9_+_109865810 | 0.04 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr1_-_169938060 | 0.03 |
ENSMUST00000027985.8
ENSMUST00000194690.6 |
Ddr2
|
discoidin domain receptor family, member 2 |
| chr13_-_59930059 | 0.03 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr1_-_136273811 | 0.03 |
ENSMUST00000048309.12
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr13_+_22534534 | 0.03 |
ENSMUST00000226909.2
ENSMUST00000227167.2 ENSMUST00000226786.2 |
Vmn1r198
|
vomeronasal 1 receptor 198 |
| chr13_+_92442488 | 0.03 |
ENSMUST00000188317.2
|
Gm20379
|
predicted gene, 20379 |
| chr13_+_83723743 | 0.03 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_80591030 | 0.03 |
ENSMUST00000105336.9
|
Dot1l
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
| chr13_+_41071077 | 0.03 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr9_-_39486831 | 0.03 |
ENSMUST00000216298.2
ENSMUST00000215194.2 |
Olfr959
|
olfactory receptor 959 |
| chr11_-_62283431 | 0.03 |
ENSMUST00000151498.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr17_-_29162794 | 0.03 |
ENSMUST00000232977.2
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr6_+_128376729 | 0.03 |
ENSMUST00000001561.12
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr2_+_120807498 | 0.03 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr7_+_23833572 | 0.03 |
ENSMUST00000205680.2
ENSMUST00000056549.9 |
Zfp235
|
zinc finger protein 235 |
| chr4_-_43584386 | 0.03 |
ENSMUST00000107884.3
|
Msmp
|
microseminoprotein, prostate associated |
| chr12_-_101924407 | 0.03 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr8_+_79235946 | 0.03 |
ENSMUST00000209490.2
ENSMUST00000034111.10 |
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr10_+_59942274 | 0.02 |
ENSMUST00000165024.3
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| chr5_+_130284431 | 0.02 |
ENSMUST00000044204.12
|
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr10_-_115197775 | 0.02 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr1_+_45350698 | 0.02 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr2_-_86061745 | 0.02 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr3_+_68491487 | 0.02 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr5_-_130284366 | 0.02 |
ENSMUST00000026387.11
|
Sbds
|
SBDS ribosome maturation factor |
| chr13_+_94954202 | 0.02 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr4_+_134042423 | 0.02 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr10_+_115653152 | 0.02 |
ENSMUST00000080630.11
ENSMUST00000179196.3 ENSMUST00000035563.15 |
Tspan8
|
tetraspanin 8 |
| chr3_-_130503041 | 0.02 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr7_+_23969822 | 0.02 |
ENSMUST00000108438.10
|
Zfp93
|
zinc finger protein 93 |
| chr1_+_51328265 | 0.02 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr12_-_83643964 | 0.02 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr12_+_25024913 | 0.02 |
ENSMUST00000066652.7
ENSMUST00000220459.2 ENSMUST00000222941.2 |
Kidins220
|
kinase D-interacting substrate 220 |
| chr1_-_43137768 | 0.02 |
ENSMUST00000190427.7
ENSMUST00000095014.8 |
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr4_+_108704982 | 0.02 |
ENSMUST00000102738.4
|
Kti12
|
KTI12 homolog, chromatin associated |
| chr2_-_6726998 | 0.02 |
ENSMUST00000182657.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr2_+_109522781 | 0.02 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr2_+_76236870 | 0.02 |
ENSMUST00000077972.11
ENSMUST00000111929.8 ENSMUST00000111930.9 |
Osbpl6
|
oxysterol binding protein-like 6 |
| chr9_+_38259893 | 0.02 |
ENSMUST00000216304.2
|
Olfr898
|
olfactory receptor 898 |
| chr2_-_181223787 | 0.02 |
ENSMUST00000057816.15
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr2_-_7400690 | 0.02 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr7_-_133384449 | 0.01 |
ENSMUST00000063669.8
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr9_+_38259707 | 0.01 |
ENSMUST00000217063.2
|
Olfr898
|
olfactory receptor 898 |
| chr8_+_47439916 | 0.01 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr6_+_58573656 | 0.01 |
ENSMUST00000031822.13
|
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr2_-_37537224 | 0.01 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr18_+_37827413 | 0.01 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr2_+_87853118 | 0.01 |
ENSMUST00000214438.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr10_+_42736771 | 0.01 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr19_-_8700728 | 0.01 |
ENSMUST00000170157.8
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chrX_+_162694397 | 0.01 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr12_-_85317359 | 0.01 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr11_-_99383938 | 0.01 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr10_+_87926932 | 0.01 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr10_-_116385007 | 0.01 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr14_-_50380137 | 0.01 |
ENSMUST00000213685.2
|
Olfr728
|
olfactory receptor 728 |
| chrX_-_140979913 | 0.01 |
ENSMUST00000042530.4
|
Gucy2f
|
guanylate cyclase 2f |
| chr2_+_89708781 | 0.01 |
ENSMUST00000111519.3
|
Olfr1257
|
olfactory receptor 1257 |
| chr12_+_69954506 | 0.01 |
ENSMUST00000223456.2
|
Atl1
|
atlastin GTPase 1 |
| chr19_-_45619559 | 0.01 |
ENSMUST00000160718.9
|
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr3_-_57209357 | 0.01 |
ENSMUST00000196979.5
ENSMUST00000029376.13 |
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr4_+_34893772 | 0.01 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr6_-_130258891 | 0.01 |
ENSMUST00000112020.2
|
Klra10
|
killer cell lectin-like receptor subfamily A, member 10 |
| chr2_-_86059313 | 0.01 |
ENSMUST00000099901.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr17_-_34337446 | 0.01 |
ENSMUST00000095347.13
|
Brd2
|
bromodomain containing 2 |
| chr4_-_45532470 | 0.01 |
ENSMUST00000147448.2
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr18_-_3280999 | 0.01 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr16_+_43184191 | 0.01 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_-_51039112 | 0.01 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr1_+_156666485 | 0.01 |
ENSMUST00000111720.2
|
Angptl1
|
angiopoietin-like 1 |
| chr4_+_97660971 | 0.01 |
ENSMUST00000152023.8
|
Nfia
|
nuclear factor I/A |
| chr2_+_110551685 | 0.01 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr12_+_71936500 | 0.01 |
ENSMUST00000221317.2
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr18_+_37580692 | 0.01 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr12_-_4957212 | 0.01 |
ENSMUST00000142867.8
|
Ubxn2a
|
UBX domain protein 2A |
| chrX_+_94942639 | 0.01 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr1_-_82746169 | 0.01 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr11_+_96920956 | 0.00 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr6_-_23655130 | 0.00 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr2_+_110551927 | 0.00 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr6_+_42241916 | 0.00 |
ENSMUST00000031895.13
|
Casp2
|
caspase 2 |
| chr5_+_4073343 | 0.00 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr1_-_82748964 | 0.00 |
ENSMUST00000223838.2
|
Gm47791
|
predicted gene, 47791 |
| chr5_+_104350475 | 0.00 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr11_-_99689758 | 0.00 |
ENSMUST00000105057.2
|
Gm11569
|
predicted gene 11569 |
| chr10_-_107321938 | 0.00 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr3_+_20011251 | 0.00 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr1_+_24717722 | 0.00 |
ENSMUST00000186096.7
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr18_-_60881679 | 0.00 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr5_-_73787646 | 0.00 |
ENSMUST00000152215.3
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr15_-_12549350 | 0.00 |
ENSMUST00000190929.2
|
Pdzd2
|
PDZ domain containing 2 |
| chr4_+_107171547 | 0.00 |
ENSMUST00000075693.12
ENSMUST00000139527.8 |
Yipf1
|
Yip1 domain family, member 1 |
| chr2_-_88963396 | 0.00 |
ENSMUST00000214027.2
ENSMUST00000215816.3 |
Olfr1222
|
olfactory receptor 1222 |
| chr13_-_67442285 | 0.00 |
ENSMUST00000224325.2
|
Zfp457
|
zinc finger protein 457 |
| chr19_+_23881821 | 0.00 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chrX_-_74423647 | 0.00 |
ENSMUST00000114085.9
|
F8
|
coagulation factor VIII |
| chr14_+_50630215 | 0.00 |
ENSMUST00000058965.4
|
Olfr736
|
olfactory receptor 736 |
| chr18_+_37622518 | 0.00 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr2_+_110551976 | 0.00 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxq1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072755 | cellular response to benomyl(GO:0072755) response to benomyl(GO:1901561) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0046725 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:2000096 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |