Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
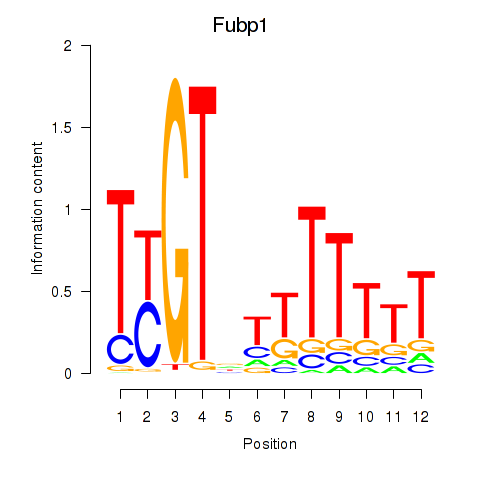
Results for Fubp1
Z-value: 0.66
Transcription factors associated with Fubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fubp1
|
ENSMUSG00000028034.16 | far upstream element (FUSE) binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Fubp1 | mm39_v1_chr3_+_151916059_151916102 | 0.28 | 6.5e-01 | Click! |
Activity profile of Fubp1 motif
Sorted Z-values of Fubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_103799873 | 0.51 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chrX_+_93679671 | 0.43 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr12_+_55431007 | 0.39 |
ENSMUST00000163070.8
|
Psma6
|
proteasome subunit alpha 6 |
| chr2_+_103800459 | 0.38 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr17_+_34423054 | 0.37 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_40586029 | 0.36 |
ENSMUST00000101347.10
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr10_+_53472853 | 0.32 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr11_-_68864666 | 0.31 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr2_+_103800553 | 0.31 |
ENSMUST00000111140.3
ENSMUST00000111139.3 |
Lmo2
|
LIM domain only 2 |
| chr12_+_17316546 | 0.28 |
ENSMUST00000057288.7
ENSMUST00000239402.2 |
Pdia6
|
protein disulfide isomerase associated 6 |
| chr18_-_43610829 | 0.28 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr4_+_144619696 | 0.28 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_-_43317017 | 0.27 |
ENSMUST00000101340.11
ENSMUST00000118368.8 ENSMUST00000020685.10 ENSMUST00000020687.15 |
Pttg1
|
pituitary tumor-transforming gene 1 |
| chr12_+_70986711 | 0.27 |
ENSMUST00000223549.2
|
Actr10
|
ARP10 actin-related protein 10 |
| chr3_-_146487102 | 0.26 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chrX_-_50106844 | 0.26 |
ENSMUST00000053593.8
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr5_-_124492734 | 0.26 |
ENSMUST00000031341.11
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr10_+_53473032 | 0.26 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr17_+_27000034 | 0.25 |
ENSMUST00000015725.16
ENSMUST00000135824.8 ENSMUST00000137989.2 |
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr11_-_69572896 | 0.23 |
ENSMUST00000066760.8
|
Senp3
|
SUMO/sentrin specific peptidase 3 |
| chr6_+_83142902 | 0.22 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr16_-_45975440 | 0.22 |
ENSMUST00000059524.7
|
Gm4737
|
predicted gene 4737 |
| chr8_-_57940834 | 0.21 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr11_-_17161504 | 0.20 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr11_-_106605772 | 0.20 |
ENSMUST00000124958.3
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr15_-_98851566 | 0.18 |
ENSMUST00000097014.7
|
Tuba1a
|
tubulin, alpha 1A |
| chr9_-_48391838 | 0.18 |
ENSMUST00000216470.2
ENSMUST00000217037.2 ENSMUST00000034524.5 ENSMUST00000213895.2 |
Rexo2
|
RNA exonuclease 2 |
| chr9_-_117979147 | 0.18 |
ENSMUST00000215799.2
ENSMUST00000044220.11 |
Cmc1
|
COX assembly mitochondrial protein 1 |
| chr3_+_89366425 | 0.17 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr8_+_84724130 | 0.17 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr1_+_179788037 | 0.16 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_+_122717852 | 0.16 |
ENSMUST00000106429.6
|
1810037I17Rik
|
RIKEN cDNA 1810037I17 gene |
| chr6_+_113366207 | 0.16 |
ENSMUST00000204026.3
|
Ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chrX_-_47602395 | 0.15 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr17_-_27423438 | 0.15 |
ENSMUST00000055117.9
|
Lemd2
|
LEM domain containing 2 |
| chr9_-_103242096 | 0.15 |
ENSMUST00000116517.9
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr11_+_4845328 | 0.15 |
ENSMUST00000038237.8
|
Thoc5
|
THO complex 5 |
| chr1_-_138103021 | 0.14 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr18_-_32044877 | 0.14 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chrX_+_149377416 | 0.14 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr11_+_4845314 | 0.14 |
ENSMUST00000101615.9
|
Thoc5
|
THO complex 5 |
| chr19_+_44551280 | 0.13 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr13_+_108452866 | 0.13 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chr9_-_103079312 | 0.12 |
ENSMUST00000035157.10
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr3_+_85946145 | 0.12 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr2_-_103609703 | 0.12 |
ENSMUST00000143188.2
|
Caprin1
|
cell cycle associated protein 1 |
| chr1_+_156193607 | 0.12 |
ENSMUST00000102782.4
|
Gm2000
|
predicted gene 2000 |
| chr2_+_104420798 | 0.12 |
ENSMUST00000028599.8
|
Cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr1_-_97904958 | 0.11 |
ENSMUST00000161567.8
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_-_146157711 | 0.11 |
ENSMUST00000169407.9
ENSMUST00000161331.8 ENSMUST00000159074.3 ENSMUST00000067837.10 |
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr11_-_106606076 | 0.11 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr2_-_93988229 | 0.11 |
ENSMUST00000028619.5
|
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr1_+_157334298 | 0.11 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334347 | 0.11 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_179788675 | 0.11 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr5_-_110987604 | 0.10 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr6_+_30401864 | 0.10 |
ENSMUST00000068240.13
ENSMUST00000068259.10 ENSMUST00000132581.8 |
Klhdc10
|
kelch domain containing 10 |
| chr16_+_17093941 | 0.10 |
ENSMUST00000164950.11
|
Tmem191c
|
transmembrane protein 191C |
| chr4_+_118266582 | 0.10 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr18_-_77855446 | 0.10 |
ENSMUST00000048192.9
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr2_+_173918715 | 0.10 |
ENSMUST00000087908.10
ENSMUST00000044638.13 ENSMUST00000156054.2 |
Stx16
|
syntaxin 16 |
| chr13_-_19521337 | 0.10 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr15_-_57982705 | 0.09 |
ENSMUST00000228783.2
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr17_+_26471870 | 0.09 |
ENSMUST00000025023.15
|
Luc7l
|
Luc7-like |
| chr5_-_121523670 | 0.09 |
ENSMUST00000146185.2
ENSMUST00000042312.14 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr10_-_59277570 | 0.09 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr1_-_138102972 | 0.09 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr3_+_96537484 | 0.09 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr19_-_56536443 | 0.09 |
ENSMUST00000182059.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr11_+_117123107 | 0.09 |
ENSMUST00000106354.9
|
Septin9
|
septin 9 |
| chr18_-_34884555 | 0.08 |
ENSMUST00000060710.9
|
Cdc25c
|
cell division cycle 25C |
| chr7_-_79974166 | 0.08 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr6_+_129510117 | 0.08 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr13_+_96524802 | 0.08 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr3_+_84859453 | 0.08 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr7_+_89053562 | 0.08 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr8_-_61407760 | 0.07 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr16_-_94171533 | 0.07 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_+_48704121 | 0.07 |
ENSMUST00000063886.4
|
Acvr2a
|
activin receptor IIA |
| chr11_-_107238956 | 0.07 |
ENSMUST00000134763.2
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr17_-_46343291 | 0.07 |
ENSMUST00000071648.12
ENSMUST00000142351.9 |
Vegfa
|
vascular endothelial growth factor A |
| chr9_+_106699073 | 0.07 |
ENSMUST00000159645.8
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr17_+_26471889 | 0.07 |
ENSMUST00000114976.9
ENSMUST00000140427.8 ENSMUST00000119928.8 |
Luc7l
|
Luc7-like |
| chr17_+_35268942 | 0.07 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr3_-_108934916 | 0.07 |
ENSMUST00000171143.2
|
Fam102b
|
family with sequence similarity 102, member B |
| chr4_-_132649798 | 0.07 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr10_+_94412116 | 0.06 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr9_+_108673171 | 0.06 |
ENSMUST00000195514.6
ENSMUST00000085018.6 ENSMUST00000192028.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr1_+_36550948 | 0.05 |
ENSMUST00000001166.14
ENSMUST00000097776.4 |
Cnnm3
|
cyclin M3 |
| chr4_+_118266526 | 0.05 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr3_+_96537235 | 0.05 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr12_-_87519032 | 0.05 |
ENSMUST00000021428.9
|
Snw1
|
SNW domain containing 1 |
| chr11_+_54517164 | 0.05 |
ENSMUST00000239168.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr12_-_32258331 | 0.05 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr16_+_8288627 | 0.05 |
ENSMUST00000046470.16
ENSMUST00000150790.2 ENSMUST00000142899.2 |
Mettl22
|
methyltransferase like 22 |
| chr6_+_35154545 | 0.05 |
ENSMUST00000170234.2
|
Nup205
|
nucleoporin 205 |
| chr14_-_24537067 | 0.05 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr2_-_77349909 | 0.05 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr1_+_60948149 | 0.05 |
ENSMUST00000027164.9
|
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr10_-_127504416 | 0.04 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr6_-_114898739 | 0.04 |
ENSMUST00000032459.14
|
Vgll4
|
vestigial like family member 4 |
| chr19_+_26728339 | 0.04 |
ENSMUST00000175953.3
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_-_56536646 | 0.04 |
ENSMUST00000182276.2
|
Dclre1a
|
DNA cross-link repair 1A |
| chr13_-_78347876 | 0.04 |
ENSMUST00000091458.13
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr18_-_84104507 | 0.04 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr12_-_32258469 | 0.04 |
ENSMUST00000085469.6
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr9_-_117081518 | 0.04 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr16_-_94171556 | 0.04 |
ENSMUST00000113906.9
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr3_-_100591906 | 0.04 |
ENSMUST00000130066.2
|
Man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr10_+_39296005 | 0.03 |
ENSMUST00000157009.8
|
Fyn
|
Fyn proto-oncogene |
| chr4_-_130169006 | 0.03 |
ENSMUST00000122374.8
|
Serinc2
|
serine incorporator 2 |
| chr14_-_72840373 | 0.03 |
ENSMUST00000162825.8
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr11_-_78074377 | 0.03 |
ENSMUST00000102483.5
|
Rpl23a
|
ribosomal protein L23A |
| chr2_-_4656870 | 0.03 |
ENSMUST00000192470.2
ENSMUST00000035721.14 ENSMUST00000152362.7 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr13_-_113317431 | 0.03 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chr7_-_110681402 | 0.03 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr2_-_164231015 | 0.03 |
ENSMUST00000167427.2
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr6_-_113354668 | 0.03 |
ENSMUST00000193384.2
|
Tada3
|
transcriptional adaptor 3 |
| chr11_+_99748741 | 0.03 |
ENSMUST00000107434.2
|
Gm11568
|
predicted gene 11568 |
| chr13_-_103901010 | 0.03 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr5_+_110988095 | 0.03 |
ENSMUST00000198373.2
|
Chek2
|
checkpoint kinase 2 |
| chr5_+_110987839 | 0.03 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr12_+_21366386 | 0.03 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr18_-_84104574 | 0.03 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr14_+_61836944 | 0.02 |
ENSMUST00000039562.8
|
Trim13
|
tripartite motif-containing 13 |
| chr2_-_85992176 | 0.02 |
ENSMUST00000099908.6
ENSMUST00000215624.2 |
Olfr1042
|
olfactory receptor 1042 |
| chr16_-_92622659 | 0.02 |
ENSMUST00000186296.2
|
Runx1
|
runt related transcription factor 1 |
| chr5_-_73789764 | 0.02 |
ENSMUST00000087177.4
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr3_+_137046828 | 0.02 |
ENSMUST00000122064.8
ENSMUST00000119475.6 |
Emcn
|
endomucin |
| chr6_+_134807097 | 0.02 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr19_-_11301919 | 0.02 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr12_+_103564479 | 0.02 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_+_65676176 | 0.02 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr4_-_147726953 | 0.02 |
ENSMUST00000133006.2
ENSMUST00000037565.14 ENSMUST00000105720.8 |
Zfp979
|
zinc finger protein 979 |
| chr11_-_99742434 | 0.02 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr7_-_103406540 | 0.02 |
ENSMUST00000106880.3
|
Olfr630
|
olfactory receptor 630 |
| chr5_+_67125759 | 0.02 |
ENSMUST00000238993.2
ENSMUST00000038188.14 |
Limch1
|
LIM and calponin homology domains 1 |
| chrX_+_162694397 | 0.02 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr3_+_59637203 | 0.02 |
ENSMUST00000168156.3
|
Aadacl2fm2
|
AADACL2 family member 2 |
| chr8_-_41668182 | 0.01 |
ENSMUST00000034003.5
|
Fgl1
|
fibrinogen-like protein 1 |
| chr2_+_65676111 | 0.01 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr5_+_88523967 | 0.01 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr4_+_146599377 | 0.01 |
ENSMUST00000140089.7
ENSMUST00000179175.3 |
Zfp981
|
zinc finger protein 981 |
| chr19_+_13610308 | 0.01 |
ENSMUST00000053113.5
|
Olfr1489
|
olfactory receptor 1489 |
| chr12_+_87921198 | 0.01 |
ENSMUST00000110145.12
ENSMUST00000181843.2 ENSMUST00000180706.8 ENSMUST00000181394.8 ENSMUST00000181326.8 ENSMUST00000181300.2 |
Gm2042
|
predicted gene 2042 |
| chr4_-_49549489 | 0.01 |
ENSMUST00000029987.10
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chr8_+_94537910 | 0.01 |
ENSMUST00000138659.9
|
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr11_+_67167950 | 0.01 |
ENSMUST00000019625.12
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal |
| chr7_-_106491314 | 0.01 |
ENSMUST00000088687.3
|
Olfr707
|
olfactory receptor 707 |
| chr4_+_62398262 | 0.01 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr2_+_111209312 | 0.01 |
ENSMUST00000209096.2
|
Olfr1284
|
olfactory receptor 1284 |
| chr12_+_35975619 | 0.01 |
ENSMUST00000042101.5
ENSMUST00000154042.2 |
Agr3
|
anterior gradient 3 |
| chr5_-_44956932 | 0.01 |
ENSMUST00000199261.2
ENSMUST00000199534.5 |
Ldb2
|
LIM domain binding 2 |
| chr5_-_82271183 | 0.01 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr4_+_147445744 | 0.01 |
ENSMUST00000133078.8
ENSMUST00000154154.2 |
Zfp978
|
zinc finger protein 978 |
| chr11_+_73819052 | 0.01 |
ENSMUST00000205791.2
|
Olfr396-ps1
|
olfactory receptor 396, pseudogene 1 |
| chr19_-_12868022 | 0.01 |
ENSMUST00000081236.3
|
Olfr1446
|
olfactory receptor 1446 |
| chr10_+_126911134 | 0.01 |
ENSMUST00000239120.2
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr7_+_107702486 | 0.01 |
ENSMUST00000104917.4
|
Olfr483
|
olfactory receptor 483 |
| chrX_-_35755483 | 0.01 |
ENSMUST00000169499.2
|
Gm14569
|
predicted gene 14569 |
| chr15_-_5093222 | 0.01 |
ENSMUST00000110689.5
|
C7
|
complement component 7 |
| chrX_-_58612709 | 0.01 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr15_-_37792635 | 0.01 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr5_-_44956981 | 0.01 |
ENSMUST00000070748.10
|
Ldb2
|
LIM domain binding 2 |
| chr6_+_18866308 | 0.01 |
ENSMUST00000031489.10
|
Ankrd7
|
ankyrin repeat domain 7 |
| chr14_-_62718581 | 0.01 |
ENSMUST00000186010.8
|
Gm4131
|
predicted gene 4131 |
| chr2_-_85590718 | 0.01 |
ENSMUST00000099916.2
|
Olfr1012
|
olfactory receptor 1012 |
| chr16_+_22769822 | 0.01 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr17_-_37625328 | 0.01 |
ENSMUST00000169373.2
|
Olfr102
|
olfactory receptor 102 |
| chr7_-_21035392 | 0.01 |
ENSMUST00000168398.2
|
Vmn1r126
|
vomeronasal 1 receptor 126 |
| chr17_-_37611375 | 0.01 |
ENSMUST00000058046.6
|
Olfr101
|
olfactory receptor 101 |
| chrX_-_166638057 | 0.01 |
ENSMUST00000238211.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr10_-_53506038 | 0.01 |
ENSMUST00000218549.3
|
Mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr9_+_39578501 | 0.01 |
ENSMUST00000215649.2
|
Olfr963
|
olfactory receptor 963 |
| chr3_+_138233004 | 0.01 |
ENSMUST00000196990.5
ENSMUST00000200239.5 ENSMUST00000200100.2 |
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr10_+_23875837 | 0.01 |
ENSMUST00000092658.2
|
Taar7b
|
trace amine-associated receptor 7B |
| chr10_+_112151245 | 0.01 |
ENSMUST00000218445.2
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr14_+_53782432 | 0.01 |
ENSMUST00000103651.4
|
Trav13-1
|
T cell receptor alpha variable 13-1 |
| chr13_-_78344492 | 0.01 |
ENSMUST00000125176.3
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr3_-_157630690 | 0.01 |
ENSMUST00000118539.2
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr5_-_87634665 | 0.01 |
ENSMUST00000201519.2
|
Gm43638
|
predicted gene 43638 |
| chr5_-_44957016 | 0.01 |
ENSMUST00000199256.5
|
Ldb2
|
LIM domain binding 2 |
| chr14_-_48904958 | 0.01 |
ENSMUST00000144465.8
ENSMUST00000133479.8 ENSMUST00000119070.8 ENSMUST00000226501.2 |
Otx2
|
orthodenticle homeobox 2 |
| chr5_-_69699932 | 0.01 |
ENSMUST00000202423.2
|
Yipf7
|
Yip1 domain family, member 7 |
| chr3_-_26207456 | 0.01 |
ENSMUST00000191835.2
|
Nlgn1
|
neuroligin 1 |
| chr5_-_69699965 | 0.01 |
ENSMUST00000031045.10
|
Yipf7
|
Yip1 domain family, member 7 |
| chr11_-_68277799 | 0.01 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr12_-_84031622 | 0.01 |
ENSMUST00000164935.3
|
Heatr4
|
HEAT repeat containing 4 |
| chr3_-_151960948 | 0.01 |
ENSMUST00000199423.5
ENSMUST00000198460.5 |
Nexn
|
nexilin |
| chr2_-_89502276 | 0.01 |
ENSMUST00000214639.2
ENSMUST00000214304.2 |
Olfr1251
|
olfactory receptor 1251 |
| chr4_-_120682859 | 0.01 |
ENSMUST00000134979.8
ENSMUST00000136236.8 ENSMUST00000043429.12 ENSMUST00000145658.2 |
Nfyc
|
nuclear transcription factor-Y gamma |
| chr10_+_129086405 | 0.01 |
ENSMUST00000097163.3
|
Olfr775
|
olfactory receptor 775 |
| chr7_+_98089623 | 0.01 |
ENSMUST00000206435.2
|
Gucy2d
|
guanylate cyclase 2d |
| chr15_+_102862862 | 0.01 |
ENSMUST00000001701.4
|
Hoxc11
|
homeobox C11 |
| chr16_+_22769844 | 0.01 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr4_-_60222580 | 0.01 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr4_+_94627755 | 0.00 |
ENSMUST00000071168.6
|
Tek
|
TEK receptor tyrosine kinase |
| chr4_-_61259997 | 0.00 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr9_-_117080869 | 0.00 |
ENSMUST00000172564.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr7_+_108229201 | 0.00 |
ENSMUST00000072914.3
|
Olfr508
|
olfactory receptor 508 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0002477 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.1 | 1.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 0.1 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 1.2 | GO:0070888 | bHLH transcription factor binding(GO:0043425) E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |