Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
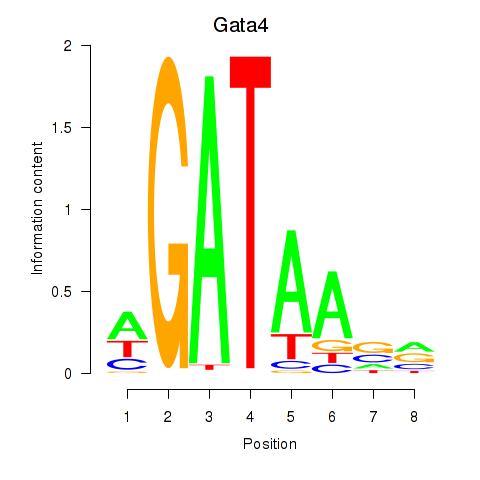
Results for Gata4
Z-value: 0.59
Transcription factors associated with Gata4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata4
|
ENSMUSG00000021944.16 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata4 | mm39_v1_chr14_-_63482668_63482714 | -0.82 | 8.6e-02 | Click! |
Activity profile of Gata4 motif
Sorted Z-values of Gata4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_84810802 | 0.50 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr4_+_102617495 | 0.41 |
ENSMUST00000072481.12
ENSMUST00000156596.8 ENSMUST00000080728.13 ENSMUST00000106882.9 |
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr6_-_129484070 | 0.36 |
ENSMUST00000183258.8
ENSMUST00000182784.4 ENSMUST00000032265.13 ENSMUST00000162815.2 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr11_+_3280401 | 0.34 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_135531548 | 0.33 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr4_-_87724533 | 0.33 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr9_-_44253630 | 0.32 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr4_+_41760454 | 0.30 |
ENSMUST00000108040.8
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr2_+_72306503 | 0.28 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr3_-_20296337 | 0.27 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr4_-_87724512 | 0.26 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr2_+_163389068 | 0.26 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr10_-_62178453 | 0.25 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr19_+_24853039 | 0.23 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr7_+_80707328 | 0.21 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chrX_-_106446928 | 0.20 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr1_+_151220222 | 0.20 |
ENSMUST00000023918.13
ENSMUST00000111887.10 ENSMUST00000097543.8 |
Ivns1abp
|
influenza virus NS1A binding protein |
| chr6_-_127086480 | 0.20 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr1_+_177272215 | 0.20 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_+_135531834 | 0.18 |
ENSMUST00000029810.6
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr8_-_46664321 | 0.18 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr10_+_61484331 | 0.18 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr1_-_46927230 | 0.18 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr6_-_115652831 | 0.18 |
ENSMUST00000112949.8
|
Raf1
|
v-raf-leukemia viral oncogene 1 |
| chr11_+_87938626 | 0.17 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr2_+_103788321 | 0.17 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr5_-_125371162 | 0.16 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr19_-_6065415 | 0.16 |
ENSMUST00000237519.2
|
Capn1
|
calpain 1 |
| chr18_-_24153363 | 0.16 |
ENSMUST00000153337.2
ENSMUST00000148525.2 |
Zfp24
|
zinc finger protein 24 |
| chr17_+_31427023 | 0.16 |
ENSMUST00000173776.2
ENSMUST00000048656.15 |
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr15_-_77037972 | 0.15 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr10_+_58207229 | 0.15 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chrX_+_111221031 | 0.15 |
ENSMUST00000026599.10
ENSMUST00000113415.2 |
Apool
|
apolipoprotein O-like |
| chr14_+_55798362 | 0.15 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr16_+_23338960 | 0.15 |
ENSMUST00000211460.2
ENSMUST00000210658.2 ENSMUST00000209198.2 ENSMUST00000210371.2 ENSMUST00000211499.2 ENSMUST00000210795.2 ENSMUST00000209422.2 |
Gm45338
Rtp4
|
predicted gene 45338 receptor transporter protein 4 |
| chr18_-_61840654 | 0.14 |
ENSMUST00000025472.7
|
Pcyox1l
|
prenylcysteine oxidase 1 like |
| chr19_-_6065799 | 0.14 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chr15_-_38518458 | 0.14 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr13_-_59930059 | 0.14 |
ENSMUST00000225581.2
|
Gm49354
|
predicted gene, 49354 |
| chr1_+_165616250 | 0.14 |
ENSMUST00000161971.8
ENSMUST00000187313.7 ENSMUST00000178336.8 ENSMUST00000005907.12 ENSMUST00000027849.11 |
Cd247
|
CD247 antigen |
| chr7_-_98887770 | 0.13 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr8_-_117809188 | 0.13 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr14_-_70945434 | 0.13 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr14_+_55798517 | 0.13 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr2_+_61542038 | 0.13 |
ENSMUST00000028278.14
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr3_+_89987749 | 0.12 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr1_-_179631190 | 0.12 |
ENSMUST00000027768.14
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr5_+_32616187 | 0.12 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr8_+_85428059 | 0.12 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr11_-_46280298 | 0.12 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr5_-_73349191 | 0.12 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr13_-_8921732 | 0.12 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr4_+_14864076 | 0.11 |
ENSMUST00000029875.4
|
Pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr4_+_102427247 | 0.11 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_-_102360664 | 0.11 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr11_+_87938519 | 0.11 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr3_+_135531409 | 0.11 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr4_-_63779562 | 0.11 |
ENSMUST00000030047.3
|
Tnfsf8
|
tumor necrosis factor (ligand) superfamily, member 8 |
| chr11_+_87938128 | 0.11 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr2_+_109522781 | 0.10 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr11_-_46280336 | 0.10 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr1_-_171108754 | 0.10 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr19_-_43512929 | 0.10 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr8_+_85428391 | 0.10 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr7_-_126398165 | 0.09 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr6_-_71609881 | 0.09 |
ENSMUST00000065509.11
ENSMUST00000207023.2 |
Kdm3a
|
lysine (K)-specific demethylase 3A |
| chr1_+_45350698 | 0.09 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr17_+_71511642 | 0.09 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr15_-_83316995 | 0.08 |
ENSMUST00000165095.9
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr7_+_51537645 | 0.08 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr2_-_23938869 | 0.08 |
ENSMUST00000114497.2
|
Hnmt
|
histamine N-methyltransferase |
| chr15_-_83054698 | 0.08 |
ENSMUST00000162178.8
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr10_+_39009951 | 0.08 |
ENSMUST00000019991.8
|
Tube1
|
tubulin, epsilon 1 |
| chr15_-_83317020 | 0.08 |
ENSMUST00000231184.2
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_6065181 | 0.08 |
ENSMUST00000236537.2
ENSMUST00000025891.11 |
Capn1
|
calpain 1 |
| chrX_+_92718695 | 0.08 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr2_+_90948481 | 0.07 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_66893280 | 0.07 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr17_+_84990541 | 0.07 |
ENSMUST00000045714.15
ENSMUST00000171915.2 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr3_+_88461059 | 0.07 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr6_+_115398996 | 0.06 |
ENSMUST00000000450.5
|
Pparg
|
peroxisome proliferator activated receptor gamma |
| chr11_+_97917746 | 0.06 |
ENSMUST00000017548.7
|
Rpl19
|
ribosomal protein L19 |
| chrX_+_149981074 | 0.06 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr9_-_44253588 | 0.06 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr14_-_54648057 | 0.06 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr2_-_86109346 | 0.06 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr7_-_98829474 | 0.06 |
ENSMUST00000207611.2
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chrX_+_92698469 | 0.06 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr13_+_107083613 | 0.06 |
ENSMUST00000022203.10
|
Dimt1
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr1_+_165616315 | 0.05 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr2_-_52448552 | 0.05 |
ENSMUST00000102760.10
ENSMUST00000102761.9 |
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_+_180678677 | 0.05 |
ENSMUST00000038091.8
|
Sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr13_+_56850795 | 0.05 |
ENSMUST00000069557.14
ENSMUST00000109876.8 |
Smad5
|
SMAD family member 5 |
| chr2_-_17465410 | 0.05 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr12_-_91815855 | 0.05 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr15_-_77037756 | 0.05 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr14_-_70585874 | 0.05 |
ENSMUST00000152067.8
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr15_-_77037926 | 0.05 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr16_-_95260104 | 0.04 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr3_+_69914946 | 0.04 |
ENSMUST00000053013.6
|
Otol1
|
otolin 1 |
| chr14_+_59716265 | 0.04 |
ENSMUST00000224893.2
|
Cab39l
|
calcium binding protein 39-like |
| chr11_-_115967873 | 0.04 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr11_+_72938609 | 0.04 |
ENSMUST00000021135.5
|
Ncbp3
|
nuclear cap binding subunit 3 |
| chr2_+_15531281 | 0.04 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr17_+_3532455 | 0.04 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr10_+_116013256 | 0.04 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr7_+_19197192 | 0.03 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr10_-_18887701 | 0.03 |
ENSMUST00000105527.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr8_+_46080840 | 0.03 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr15_+_41615084 | 0.03 |
ENSMUST00000229511.2
ENSMUST00000229836.2 |
Oxr1
|
oxidation resistance 1 |
| chrX_+_55825033 | 0.03 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chrX_-_7439082 | 0.03 |
ENSMUST00000132788.2
|
Ppp1r3f
|
protein phosphatase 1, regulatory subunit 3F |
| chr12_+_103564479 | 0.03 |
ENSMUST00000190151.2
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_+_85805557 | 0.03 |
ENSMUST00000082191.5
|
Olfr1029
|
olfactory receptor 1029 |
| chr10_-_59452489 | 0.03 |
ENSMUST00000020312.13
|
Mcu
|
mitochondrial calcium uniporter |
| chr1_-_159981132 | 0.02 |
ENSMUST00000039178.12
|
Tnn
|
tenascin N |
| chr11_+_4959515 | 0.02 |
ENSMUST00000101613.3
|
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr17_-_15596230 | 0.02 |
ENSMUST00000014917.8
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr4_-_14621805 | 0.02 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_+_10180716 | 0.02 |
ENSMUST00000055964.8
|
Vmn1r67
|
vomeronasal 1 receptor 67 |
| chr4_+_102112189 | 0.02 |
ENSMUST00000106908.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr4_-_110144676 | 0.02 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr3_+_66893031 | 0.02 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr7_-_133304244 | 0.02 |
ENSMUST00000209636.2
ENSMUST00000153698.3 |
Uros
|
uroporphyrinogen III synthase |
| chrX_-_59449137 | 0.02 |
ENSMUST00000033480.13
ENSMUST00000101527.3 |
Atp11c
|
ATPase, class VI, type 11C |
| chr17_-_33932346 | 0.02 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr3_-_63758672 | 0.02 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr2_+_65676176 | 0.02 |
ENSMUST00000053910.10
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr12_+_95658987 | 0.02 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_+_132844971 | 0.02 |
ENSMUST00000082014.2
|
Tas2r110
|
taste receptor, type 2, member 110 |
| chr3_-_59170245 | 0.02 |
ENSMUST00000050360.14
ENSMUST00000199609.2 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr15_-_103159892 | 0.02 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr10_-_80269436 | 0.02 |
ENSMUST00000105346.10
ENSMUST00000020377.13 ENSMUST00000105340.8 ENSMUST00000020379.13 ENSMUST00000105344.8 ENSMUST00000105342.8 ENSMUST00000105345.10 ENSMUST00000105343.8 |
Tcf3
|
transcription factor 3 |
| chr4_-_32923505 | 0.01 |
ENSMUST00000084749.8
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr17_+_20722561 | 0.01 |
ENSMUST00000061660.2
|
Vmn1r225
|
vomeronasal 1 receptor 225 |
| chrX_+_55824797 | 0.01 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr12_+_31488208 | 0.01 |
ENSMUST00000001254.6
|
Slc26a3
|
solute carrier family 26, member 3 |
| chr16_-_44379226 | 0.01 |
ENSMUST00000114634.3
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| chr10_+_69055215 | 0.01 |
ENSMUST00000172261.3
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr5_-_83502966 | 0.01 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr14_-_50322136 | 0.01 |
ENSMUST00000072370.3
|
Olfr726
|
olfactory receptor 726 |
| chr1_-_57008986 | 0.01 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr17_-_84495364 | 0.01 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr10_-_129010923 | 0.01 |
ENSMUST00000169800.4
|
Olfr772
|
olfactory receptor 772 |
| chr14_-_68771138 | 0.01 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr7_-_4633186 | 0.01 |
ENSMUST00000205360.2
ENSMUST00000206610.2 |
Tmem86b
|
transmembrane protein 86B |
| chr6_-_146988499 | 0.01 |
ENSMUST00000123367.2
ENSMUST00000100780.3 |
Mansc4
|
MANSC domain containing 4 |
| chr3_+_29568055 | 0.01 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr18_+_37433852 | 0.01 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr6_+_129374441 | 0.01 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr1_+_12788720 | 0.01 |
ENSMUST00000088585.10
|
Sulf1
|
sulfatase 1 |
| chr14_+_53157900 | 0.01 |
ENSMUST00000178252.3
|
Trav9d-3
|
T cell receptor alpha variable 9D-3 |
| chr3_+_79792238 | 0.01 |
ENSMUST00000135021.2
|
Gask1b
|
golgi associated kinase 1B |
| chr2_+_14878480 | 0.01 |
ENSMUST00000114719.7
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr5_-_24534554 | 0.01 |
ENSMUST00000115098.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr6_+_41279199 | 0.01 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr4_-_32923389 | 0.01 |
ENSMUST00000035719.11
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr9_+_21176582 | 0.01 |
ENSMUST00000065005.5
|
Atg4d
|
autophagy related 4D, cysteine peptidase |
| chr14_-_54647647 | 0.01 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr4_-_14621497 | 0.01 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr16_-_23339329 | 0.01 |
ENSMUST00000230040.2
ENSMUST00000229619.2 |
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr19_+_27299939 | 0.01 |
ENSMUST00000056708.4
|
Kcnv2
|
potassium channel, subfamily V, member 2 |
| chrX_+_10118544 | 0.01 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr3_+_108272205 | 0.01 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chr16_-_59022882 | 0.01 |
ENSMUST00000064452.3
|
Olfr198
|
olfactory receptor 198 |
| chr19_-_46661501 | 0.01 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr4_+_102111936 | 0.01 |
ENSMUST00000106907.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr14_-_55204054 | 0.01 |
ENSMUST00000226297.2
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr10_-_116732813 | 0.01 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr3_-_75177378 | 0.01 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr7_+_45063079 | 0.01 |
ENSMUST00000058879.8
|
Ntf5
|
neurotrophin 5 |
| chr14_+_53468890 | 0.01 |
ENSMUST00000177705.3
|
Trav9n-3
|
T cell receptor alpha variable 9N-3 |
| chr19_-_40175709 | 0.01 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr10_-_128974829 | 0.01 |
ENSMUST00000203887.3
ENSMUST00000204250.3 ENSMUST00000204712.4 |
Olfr770
|
olfactory receptor 770 |
| chr14_-_55204092 | 0.01 |
ENSMUST00000081857.14
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr5_-_83502793 | 0.01 |
ENSMUST00000146669.2
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr16_-_34083549 | 0.01 |
ENSMUST00000114949.8
ENSMUST00000114954.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr9_-_40039335 | 0.01 |
ENSMUST00000060345.6
|
Olfr985
|
olfactory receptor 985 |
| chr13_-_56696310 | 0.01 |
ENSMUST00000062806.6
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr13_-_56696222 | 0.01 |
ENSMUST00000225183.2
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_93184854 | 0.01 |
ENSMUST00000180308.3
|
Flg
|
filaggrin |
| chr12_+_78243846 | 0.01 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr3_+_89043440 | 0.01 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr7_-_135130374 | 0.01 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr2_+_65676111 | 0.01 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr7_-_30555592 | 0.01 |
ENSMUST00000185748.2
ENSMUST00000094583.2 |
Ffar3
|
free fatty acid receptor 3 |
| chr14_+_52680447 | 0.01 |
ENSMUST00000205900.3
ENSMUST00000215030.2 ENSMUST00000206100.2 ENSMUST00000206718.2 |
Olfr1509
|
olfactory receptor 1509 |
| chr7_+_102526329 | 0.01 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr5_+_104318542 | 0.01 |
ENSMUST00000112771.2
|
Dspp
|
dentin sialophosphoprotein |
| chr13_-_21327251 | 0.01 |
ENSMUST00000055298.6
|
Olfr1368
|
olfactory receptor 1368 |
| chr7_+_123061535 | 0.01 |
ENSMUST00000098056.6
|
Aqp8
|
aquaporin 8 |
| chr19_-_10655391 | 0.01 |
ENSMUST00000025647.7
|
Pga5
|
pepsinogen 5, group I |
| chr2_-_25124240 | 0.01 |
ENSMUST00000006638.8
|
Slc34a3
|
solute carrier family 34 (sodium phosphate), member 3 |
| chr7_+_119499322 | 0.01 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr5_-_82271183 | 0.01 |
ENSMUST00000186079.2
ENSMUST00000185607.2 |
1700031L13Rik
|
RIKEN cDNA 1700031L13 gene |
| chr2_+_74534959 | 0.01 |
ENSMUST00000151380.2
|
Hoxd8
|
homeobox D8 |
| chr9_+_38384041 | 0.01 |
ENSMUST00000051111.3
|
Olfr905
|
olfactory receptor 905 |
| chr17_+_38378064 | 0.01 |
ENSMUST00000087129.3
|
Olfr130
|
olfactory receptor 130 |
| chr11_-_99213769 | 0.01 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr19_-_11058452 | 0.01 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.3 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.1 | 0.2 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.1 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.6 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.3 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0002632 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.3 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |