Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
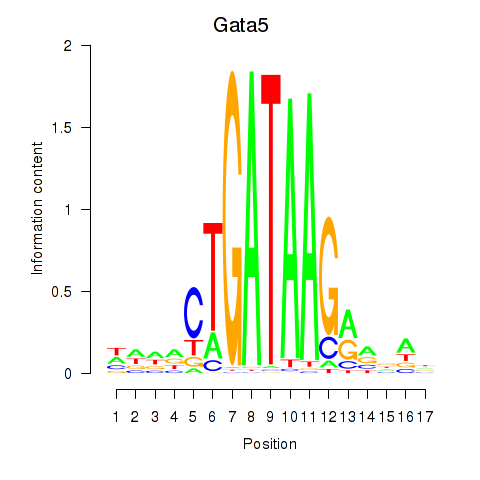
Results for Gata5
Z-value: 0.60
Transcription factors associated with Gata5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata5
|
ENSMUSG00000015627.6 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata5 | mm39_v1_chr2_-_179976458_179976509 | -0.14 | 8.2e-01 | Click! |
Activity profile of Gata5 motif
Sorted Z-values of Gata5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_103477126 | 0.65 |
ENSMUST00000023934.8
|
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr11_+_32233511 | 0.54 |
ENSMUST00000093209.4
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr17_-_34218301 | 0.49 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr11_+_32246489 | 0.45 |
ENSMUST00000093207.4
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr3_-_94794256 | 0.34 |
ENSMUST00000005923.7
|
Psmb4
|
proteasome (prosome, macropain) subunit, beta type 4 |
| chr1_-_131025562 | 0.29 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr19_-_32038838 | 0.28 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr2_-_153474599 | 0.26 |
ENSMUST00000109782.2
|
Commd7
|
COMM domain containing 7 |
| chr7_-_103928939 | 0.26 |
ENSMUST00000051795.10
|
Trim5
|
tripartite motif-containing 5 |
| chr11_+_96920751 | 0.25 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr6_-_127086480 | 0.25 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr2_+_72306503 | 0.25 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr16_-_33916354 | 0.24 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr17_+_41121979 | 0.22 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chrX_-_8059597 | 0.19 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chrX_+_52076998 | 0.18 |
ENSMUST00000026723.9
|
Hprt
|
hypoxanthine guanine phosphoribosyl transferase |
| chr2_-_25114660 | 0.18 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr17_+_19724994 | 0.17 |
ENSMUST00000166081.3
ENSMUST00000231465.2 |
Vmn2r100
|
vomeronasal 2, receptor 100 |
| chr6_+_137712076 | 0.17 |
ENSMUST00000064910.7
|
Strap
|
serine/threonine kinase receptor associated protein |
| chr17_+_48653493 | 0.17 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr1_-_46927230 | 0.17 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_-_71198091 | 0.16 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr6_+_134988572 | 0.16 |
ENSMUST00000032326.11
ENSMUST00000130851.8 ENSMUST00000205244.3 ENSMUST00000205055.3 ENSMUST00000204646.3 ENSMUST00000154558.3 |
Ddx47
|
DEAD box helicase 47 |
| chr12_-_108859123 | 0.15 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr6_-_34294377 | 0.15 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr7_+_13011180 | 0.15 |
ENSMUST00000177588.10
|
Lig1
|
ligase I, DNA, ATP-dependent |
| chr15_-_38518458 | 0.15 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr12_-_103392039 | 0.15 |
ENSMUST00000110001.4
ENSMUST00000223233.2 ENSMUST00000044923.15 ENSMUST00000221211.2 |
Ddx24
|
DEAD box helicase 24 |
| chr5_-_35897331 | 0.14 |
ENSMUST00000201511.2
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr15_-_77037972 | 0.14 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_-_133177638 | 0.14 |
ENSMUST00000113252.8
|
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr16_+_17328924 | 0.13 |
ENSMUST00000232372.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr19_-_43512929 | 0.13 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr19_+_24853039 | 0.13 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr7_-_98887770 | 0.13 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr2_+_26473870 | 0.12 |
ENSMUST00000100290.13
ENSMUST00000238983.2 ENSMUST00000102907.13 |
Egfl7
|
EGF-like domain 7 |
| chr2_-_153474695 | 0.12 |
ENSMUST00000071852.10
|
Commd7
|
COMM domain containing 7 |
| chr16_+_36755338 | 0.12 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr5_+_88731366 | 0.12 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr10_+_45453907 | 0.12 |
ENSMUST00000037044.13
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr4_-_59438633 | 0.12 |
ENSMUST00000040166.14
ENSMUST00000107544.2 |
Susd1
|
sushi domain containing 1 |
| chr6_-_68609426 | 0.11 |
ENSMUST00000103328.3
|
Igkv10-96
|
immunoglobulin kappa variable 10-96 |
| chr5_-_73349191 | 0.11 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr3_-_34135408 | 0.11 |
ENSMUST00000117223.4
ENSMUST00000120805.8 ENSMUST00000011029.12 ENSMUST00000108195.10 |
Dnajc19
|
DnaJ heat shock protein family (Hsp40) member C19 |
| chr15_-_89361571 | 0.11 |
ENSMUST00000165199.8
|
Arsa
|
arylsulfatase A |
| chr17_-_46956054 | 0.11 |
ENSMUST00000003642.7
|
Klc4
|
kinesin light chain 4 |
| chr16_+_49675682 | 0.11 |
ENSMUST00000114496.3
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr1_+_180678677 | 0.10 |
ENSMUST00000038091.8
|
Sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr9_+_74945101 | 0.10 |
ENSMUST00000167885.8
ENSMUST00000169188.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr4_-_150087587 | 0.10 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr3_-_52924616 | 0.10 |
ENSMUST00000193432.6
ENSMUST00000195183.2 ENSMUST00000036665.10 |
Cog6
|
component of oligomeric golgi complex 6 |
| chr13_-_62668265 | 0.10 |
ENSMUST00000082203.7
|
Zfp934
|
zinc finger protein 934 |
| chr3_-_97517472 | 0.10 |
ENSMUST00000029730.5
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr8_+_53964721 | 0.10 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr15_-_60696790 | 0.09 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr3_-_14873406 | 0.09 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr3_-_95014218 | 0.09 |
ENSMUST00000107233.9
|
Pip5k1a
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| chr18_+_46730765 | 0.09 |
ENSMUST00000238168.2
ENSMUST00000078079.11 ENSMUST00000168382.2 ENSMUST00000235849.2 ENSMUST00000235973.2 ENSMUST00000235455.2 ENSMUST00000237478.2 |
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr7_-_29772226 | 0.09 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr1_-_85248264 | 0.09 |
ENSMUST00000093506.12
ENSMUST00000064341.9 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr1_+_180770064 | 0.09 |
ENSMUST00000159436.8
|
Tmem63a
|
transmembrane protein 63a |
| chr7_-_4633472 | 0.08 |
ENSMUST00000055085.8
|
Tmem86b
|
transmembrane protein 86B |
| chr4_+_135583018 | 0.08 |
ENSMUST00000105853.10
ENSMUST00000097844.9 ENSMUST00000102544.9 ENSMUST00000126641.2 |
Srsf10
|
serine and arginine-rich splicing factor 10 |
| chr7_-_141014477 | 0.08 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr2_-_23938869 | 0.08 |
ENSMUST00000114497.2
|
Hnmt
|
histamine N-methyltransferase |
| chr11_-_87249837 | 0.08 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr15_+_102391614 | 0.08 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_+_34361153 | 0.08 |
ENSMUST00000038383.14
ENSMUST00000115051.8 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr11_-_101442663 | 0.08 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr9_-_57460004 | 0.08 |
ENSMUST00000034856.15
|
Mpi
|
mannose phosphate isomerase |
| chr2_+_85804239 | 0.08 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr6_-_69584812 | 0.08 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chrX_-_133177717 | 0.07 |
ENSMUST00000087541.12
ENSMUST00000087540.4 |
Trmt2b
|
TRM2 tRNA methyltransferase 2B |
| chr9_-_35030479 | 0.07 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chrX_-_106446928 | 0.07 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr2_-_37249247 | 0.07 |
ENSMUST00000112940.8
ENSMUST00000009174.15 |
Pdcl
|
phosducin-like |
| chr11_+_68979332 | 0.07 |
ENSMUST00000117780.2
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr4_+_15265798 | 0.07 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr13_-_62519750 | 0.07 |
ENSMUST00000107989.7
|
Gm3604
|
predicted gene 3604 |
| chr15_-_76491758 | 0.07 |
ENSMUST00000071898.7
|
Cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr15_-_76491684 | 0.07 |
ENSMUST00000230157.2
|
Cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr12_+_83567240 | 0.07 |
ENSMUST00000021645.9
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr3_-_14843512 | 0.07 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr7_+_27430823 | 0.07 |
ENSMUST00000130997.8
ENSMUST00000042641.14 |
Zfp60
|
zinc finger protein 60 |
| chr10_+_58207229 | 0.07 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_+_47605208 | 0.06 |
ENSMUST00000151405.9
|
Lgals3
|
lectin, galactose binding, soluble 3 |
| chr11_+_96920956 | 0.06 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr6_+_58810674 | 0.06 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr15_+_100052260 | 0.06 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr11_+_87744033 | 0.06 |
ENSMUST00000038196.7
|
Mks1
|
MKS transition zone complex subunit 1 |
| chr12_+_83567303 | 0.06 |
ENSMUST00000222502.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr9_+_110627219 | 0.06 |
ENSMUST00000035715.8
|
Prss42
|
protease, serine 42 |
| chr17_+_35133435 | 0.06 |
ENSMUST00000007249.15
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr12_-_112641260 | 0.06 |
ENSMUST00000144550.9
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr16_-_50151350 | 0.06 |
ENSMUST00000114488.8
|
Bbx
|
bobby sox HMG box containing |
| chr3_-_35991548 | 0.05 |
ENSMUST00000199173.3
ENSMUST00000148465.8 |
Dcun1d1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| chr9_-_102496047 | 0.05 |
ENSMUST00000215253.2
|
Cep63
|
centrosomal protein 63 |
| chr1_-_173703424 | 0.05 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr6_+_122285615 | 0.05 |
ENSMUST00000007602.15
ENSMUST00000112610.2 |
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr6_-_71609881 | 0.05 |
ENSMUST00000065509.11
ENSMUST00000207023.2 |
Kdm3a
|
lysine (K)-specific demethylase 3A |
| chr6_+_81900630 | 0.05 |
ENSMUST00000043195.11
|
Gcfc2
|
GC-rich sequence DNA binding factor 2 |
| chr2_-_150253257 | 0.05 |
ENSMUST00000185796.2
|
Zfp442
|
zinc finger protein 442 |
| chr1_+_85528392 | 0.05 |
ENSMUST00000080204.11
|
Sp140
|
Sp140 nuclear body protein |
| chr11_+_61544085 | 0.05 |
ENSMUST00000004959.3
|
Grap
|
GRB2-related adaptor protein |
| chr7_+_80707328 | 0.05 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr11_+_115656246 | 0.05 |
ENSMUST00000093912.11
ENSMUST00000136720.8 ENSMUST00000103034.10 ENSMUST00000141871.8 |
Tmem94
|
transmembrane protein 94 |
| chr4_+_103000248 | 0.05 |
ENSMUST00000106855.2
|
Mier1
|
MEIR1 treanscription regulator |
| chr2_+_36575800 | 0.05 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr4_+_149629559 | 0.04 |
ENSMUST00000105692.2
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr9_-_123461593 | 0.04 |
ENSMUST00000026273.11
|
Slc6a20b
|
solute carrier family 6 (neurotransmitter transporter), member 20B |
| chr5_+_88731386 | 0.04 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr7_-_141014192 | 0.04 |
ENSMUST00000201127.5
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr7_-_141014336 | 0.04 |
ENSMUST00000136354.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr9_-_122016671 | 0.04 |
ENSMUST00000216738.2
|
Ano10
|
anoctamin 10 |
| chr2_+_109522781 | 0.04 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr6_-_40562700 | 0.04 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr5_+_93241287 | 0.03 |
ENSMUST00000074733.11
ENSMUST00000201700.4 ENSMUST00000202196.4 ENSMUST00000202308.4 |
Septin11
|
septin 11 |
| chr16_-_50252703 | 0.03 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr9_-_36678868 | 0.03 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr13_+_108452866 | 0.03 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chr8_-_117809188 | 0.03 |
ENSMUST00000109093.9
ENSMUST00000098375.6 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr8_+_85428059 | 0.03 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr2_+_84867554 | 0.03 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr1_-_136273811 | 0.03 |
ENSMUST00000048309.12
|
Camsap2
|
calmodulin regulated spectrin-associated protein family, member 2 |
| chr13_-_67480588 | 0.03 |
ENSMUST00000109735.9
ENSMUST00000168892.9 |
Zfp595
|
zinc finger protein 595 |
| chr17_-_24746911 | 0.03 |
ENSMUST00000176652.8
|
Traf7
|
TNF receptor-associated factor 7 |
| chr1_+_192984278 | 0.02 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr11_+_116423266 | 0.02 |
ENSMUST00000106386.8
ENSMUST00000145737.8 ENSMUST00000155102.8 ENSMUST00000063446.13 |
Sphk1
|
sphingosine kinase 1 |
| chr2_+_90948481 | 0.02 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr19_-_11852453 | 0.02 |
ENSMUST00000213954.2
ENSMUST00000217617.2 |
Olfr1419
|
olfactory receptor 1419 |
| chr1_-_170755136 | 0.02 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr10_-_89568106 | 0.02 |
ENSMUST00000020109.5
|
Actr6
|
ARP6 actin-related protein 6 |
| chr15_-_77037926 | 0.02 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr14_+_70694887 | 0.02 |
ENSMUST00000003561.10
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr1_+_51328265 | 0.02 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr17_-_36207965 | 0.02 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr2_-_17465410 | 0.02 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr12_+_108859557 | 0.02 |
ENSMUST00000221377.2
|
Wdr25
|
WD repeat domain 25 |
| chr13_-_100338469 | 0.02 |
ENSMUST00000167986.3
ENSMUST00000117913.8 |
Naip2
|
NLR family, apoptosis inhibitory protein 2 |
| chrX_-_133012600 | 0.02 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr5_-_87240405 | 0.02 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr9_+_38684364 | 0.02 |
ENSMUST00000217114.3
ENSMUST00000213958.3 |
Olfr921
|
olfactory receptor 921 |
| chr15_+_80507671 | 0.02 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr16_+_58490625 | 0.01 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chr19_+_12647803 | 0.01 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr2_+_87576198 | 0.01 |
ENSMUST00000217572.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr4_+_6191084 | 0.01 |
ENSMUST00000029907.6
|
Ubxn2b
|
UBX domain protein 2B |
| chr19_-_38807600 | 0.01 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr6_+_68402550 | 0.01 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr1_+_172204109 | 0.01 |
ENSMUST00000052455.4
|
Pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr6_+_41331039 | 0.01 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr17_-_21110913 | 0.01 |
ENSMUST00000061278.2
|
Vmn1r231
|
vomeronasal 1 receptor 231 |
| chr2_-_88832634 | 0.01 |
ENSMUST00000188399.3
|
Olfr1215
|
olfactory receptor 1215 |
| chr12_+_59113659 | 0.01 |
ENSMUST00000021381.6
|
Pnn
|
pinin |
| chr7_-_141014445 | 0.01 |
ENSMUST00000133021.2
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr6_+_90279064 | 0.01 |
ENSMUST00000167550.4
|
BC048671
|
cDNA sequence BC048671 |
| chr1_-_132295617 | 0.01 |
ENSMUST00000142609.8
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr11_+_89921121 | 0.01 |
ENSMUST00000092788.4
|
Tmem100
|
transmembrane protein 100 |
| chr5_+_32616187 | 0.01 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr4_-_119047180 | 0.01 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr6_-_68887957 | 0.01 |
ENSMUST00000200454.2
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr14_-_50322136 | 0.01 |
ENSMUST00000072370.3
|
Olfr726
|
olfactory receptor 726 |
| chr8_-_83128437 | 0.01 |
ENSMUST00000209573.3
|
Il15
|
interleukin 15 |
| chr7_+_10180716 | 0.01 |
ENSMUST00000055964.8
|
Vmn1r67
|
vomeronasal 1 receptor 67 |
| chr2_+_85612365 | 0.01 |
ENSMUST00000215945.2
|
Olfr1015
|
olfactory receptor 1015 |
| chr7_+_107501834 | 0.01 |
ENSMUST00000210420.2
|
Olfr472
|
olfactory receptor 472 |
| chr11_-_99695272 | 0.01 |
ENSMUST00000105056.2
|
Gm11554
|
predicted gene 11554 |
| chr17_-_24746804 | 0.01 |
ENSMUST00000176353.8
ENSMUST00000176237.8 |
Traf7
|
TNF receptor-associated factor 7 |
| chr9_-_50439942 | 0.01 |
ENSMUST00000214873.2
ENSMUST00000034570.7 ENSMUST00000213144.2 ENSMUST00000215416.2 |
Pts
|
6-pyruvoyl-tetrahydropterin synthase |
| chr2_+_86655007 | 0.01 |
ENSMUST00000217509.2
|
Olfr1094
|
olfactory receptor 1094 |
| chr14_-_51295099 | 0.01 |
ENSMUST00000227764.2
|
Rnase12
|
ribonuclease, RNase A family, 12 (non-active) |
| chr16_-_58850641 | 0.01 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr2_+_10375304 | 0.01 |
ENSMUST00000114862.8
ENSMUST00000114861.8 |
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr2_+_85745537 | 0.01 |
ENSMUST00000213474.3
|
Olfr1025-ps1
|
olfactory receptor 1025, pseudogene 1 |
| chr6_-_3988900 | 0.01 |
ENSMUST00000183682.3
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr10_+_129910278 | 0.01 |
ENSMUST00000080460.3
|
Olfr822
|
olfactory receptor 822 |
| chr14_-_68771138 | 0.01 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr3_+_142300601 | 0.01 |
ENSMUST00000029936.5
|
Gbp2b
|
guanylate binding protein 2b |
| chr5_-_83502966 | 0.01 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr19_-_46661501 | 0.01 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr19_+_13477910 | 0.01 |
ENSMUST00000217001.2
|
Olfr1477
|
olfactory receptor 1477 |
| chrX_-_146236703 | 0.01 |
ENSMUST00000127465.3
ENSMUST00000237076.2 |
Vmn1r239-ps
|
vomeronasal 1 receptor 239, pseudogene |
| chr14_-_52273600 | 0.01 |
ENSMUST00000214342.2
|
Olfr221
|
olfactory receptor 221 |
| chr10_-_129000620 | 0.01 |
ENSMUST00000214271.2
|
Olfr771
|
olfactory receptor 771 |
| chrX_-_133012457 | 0.00 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr4_-_119047146 | 0.00 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr11_-_99556781 | 0.00 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr6_-_57956287 | 0.00 |
ENSMUST00000228585.2
|
Vmn1r25
|
vomeronasal 1 receptor 25 |
| chr6_+_78347636 | 0.00 |
ENSMUST00000204873.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr2_-_134396268 | 0.00 |
ENSMUST00000028704.3
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr13_+_19524136 | 0.00 |
ENSMUST00000103564.3
|
Trgv1
|
T cell receptor gamma, variable 1 |
| chr14_+_52904815 | 0.00 |
ENSMUST00000198058.2
ENSMUST00000103573.3 |
Trav6-2
|
T cell receptor alpha variable 6-2 |
| chr3_+_93184854 | 0.00 |
ENSMUST00000180308.3
|
Flg
|
filaggrin |
| chr2_+_87820457 | 0.00 |
ENSMUST00000102622.2
|
Olfr1158
|
olfactory receptor 1158 |
| chr6_-_3988835 | 0.00 |
ENSMUST00000203257.2
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr5_-_83502793 | 0.00 |
ENSMUST00000146669.2
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr19_-_46661321 | 0.00 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr13_+_67981349 | 0.00 |
ENSMUST00000222626.2
ENSMUST00000060609.8 |
Gm10037
|
predicted gene 10037 |
| chr19_-_12206908 | 0.00 |
ENSMUST00000180978.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr3_+_29568055 | 0.00 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.2 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 0.1 | 0.2 | GO:0046038 | GMP catabolic process(GO:0046038) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.2 | GO:0002585 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.1 | 0.3 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.5 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:2000521 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) thiamine pyrophosphate transport(GO:0030974) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0052805 | imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |