Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
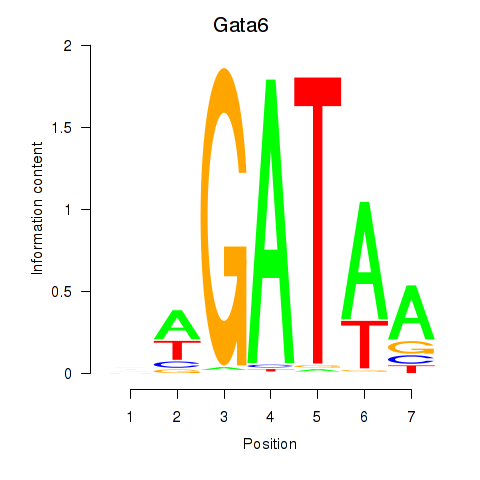
Results for Gata6
Z-value: 0.49
Transcription factors associated with Gata6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gata6
|
ENSMUSG00000005836.11 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata6 | mm39_v1_chr18_+_11052458_11052479 | 0.48 | 4.1e-01 | Click! |
Activity profile of Gata6 motif
Sorted Z-values of Gata6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_20296337 | 0.87 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr2_+_163389068 | 0.37 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr13_-_55676334 | 0.18 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr6_+_58810674 | 0.18 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr16_-_95260104 | 0.12 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr2_+_72306503 | 0.12 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr1_-_144651157 | 0.12 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr2_-_125701059 | 0.11 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr11_-_46280298 | 0.10 |
ENSMUST00000109237.9
|
Itk
|
IL2 inducible T cell kinase |
| chr15_-_103159892 | 0.10 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr9_+_62249730 | 0.10 |
ENSMUST00000156461.2
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr9_-_44253630 | 0.09 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr11_-_46280336 | 0.09 |
ENSMUST00000020664.13
|
Itk
|
IL2 inducible T cell kinase |
| chr3_-_92336712 | 0.09 |
ENSMUST00000192538.2
ENSMUST00000047300.8 |
Gm9774
|
predicted pseudogene 9774 |
| chr2_+_110427643 | 0.09 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr1_-_46927230 | 0.09 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr17_+_71511642 | 0.09 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr13_+_24986003 | 0.09 |
ENSMUST00000155575.2
|
BC005537
|
cDNA sequence BC005537 |
| chr2_+_125701054 | 0.08 |
ENSMUST00000028636.13
ENSMUST00000125084.8 |
Galk2
|
galactokinase 2 |
| chr10_+_52293617 | 0.08 |
ENSMUST00000023830.16
|
Nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr7_+_119499322 | 0.08 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr11_+_97917520 | 0.07 |
ENSMUST00000092425.11
|
Rpl19
|
ribosomal protein L19 |
| chr13_+_19398273 | 0.07 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chr2_+_143757193 | 0.07 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr17_+_34823790 | 0.06 |
ENSMUST00000173242.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr10_-_62178453 | 0.06 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr6_+_58808733 | 0.06 |
ENSMUST00000126292.8
ENSMUST00000031823.12 |
Herc3
|
hect domain and RLD 3 |
| chr11_+_97917746 | 0.05 |
ENSMUST00000017548.7
|
Rpl19
|
ribosomal protein L19 |
| chr10_+_79889322 | 0.05 |
ENSMUST00000105372.9
|
Gpx4
|
glutathione peroxidase 4 |
| chr3_+_85946145 | 0.05 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr15_-_38518458 | 0.04 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chrX_+_149981074 | 0.04 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr2_+_109522781 | 0.04 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr2_-_7400690 | 0.04 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr2_+_90948481 | 0.04 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr2_-_17465410 | 0.04 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr2_-_172212426 | 0.04 |
ENSMUST00000109139.8
ENSMUST00000028997.8 ENSMUST00000109140.10 |
Aurka
|
aurora kinase A |
| chr18_+_11052458 | 0.04 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr4_-_132649798 | 0.04 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr2_-_52225763 | 0.03 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr2_+_80447389 | 0.03 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr11_-_46280281 | 0.03 |
ENSMUST00000101306.4
|
Itk
|
IL2 inducible T cell kinase |
| chr12_+_95658987 | 0.03 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr7_-_135130374 | 0.02 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr13_-_103042554 | 0.02 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_+_46080840 | 0.02 |
ENSMUST00000135336.9
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_+_35861554 | 0.02 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr6_-_146988499 | 0.02 |
ENSMUST00000123367.2
ENSMUST00000100780.3 |
Mansc4
|
MANSC domain containing 4 |
| chr1_+_12762501 | 0.02 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr9_-_44253588 | 0.02 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr11_-_115967873 | 0.02 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr17_+_88933957 | 0.02 |
ENSMUST00000163588.8
ENSMUST00000064035.13 |
Ston1
|
stonin 1 |
| chr1_+_45350698 | 0.02 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr8_-_86091970 | 0.02 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_-_110144676 | 0.01 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr5_+_20112771 | 0.01 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr11_-_99556781 | 0.01 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr5_-_87240405 | 0.01 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr16_+_78727829 | 0.01 |
ENSMUST00000114216.2
ENSMUST00000069148.13 ENSMUST00000023568.14 |
Chodl
|
chondrolectin |
| chr3_+_59914164 | 0.01 |
ENSMUST00000169794.2
|
Aadacl2
|
arylacetamide deacetylase like 2 |
| chr5_-_30619246 | 0.01 |
ENSMUST00000114747.9
ENSMUST00000074171.10 |
Otof
|
otoferlin |
| chr14_-_7648948 | 0.01 |
ENSMUST00000112630.9
ENSMUST00000112631.9 |
Nek10
|
NIMA (never in mitosis gene a)- related kinase 10 |
| chr11_-_99383938 | 0.01 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr15_+_41615084 | 0.01 |
ENSMUST00000229511.2
ENSMUST00000229836.2 |
Oxr1
|
oxidation resistance 1 |
| chr2_-_77000878 | 0.01 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr4_-_137137088 | 0.01 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr6_-_127086480 | 0.01 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr19_-_40175709 | 0.01 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr18_+_76192529 | 0.01 |
ENSMUST00000167921.2
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr1_-_144052997 | 0.01 |
ENSMUST00000111941.2
ENSMUST00000052375.8 |
Rgs13
|
regulator of G-protein signaling 13 |
| chr14_+_33662976 | 0.01 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr15_+_57849269 | 0.01 |
ENSMUST00000050374.3
|
Fam83a
|
family with sequence similarity 83, member A |
| chr7_-_30555592 | 0.01 |
ENSMUST00000185748.2
ENSMUST00000094583.2 |
Ffar3
|
free fatty acid receptor 3 |
| chr11_-_99213769 | 0.01 |
ENSMUST00000038004.3
|
Krt25
|
keratin 25 |
| chr11_-_70111796 | 0.01 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr8_-_86091946 | 0.01 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr19_-_39801188 | 0.01 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr3_+_108272205 | 0.01 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chr1_-_172958803 | 0.01 |
ENSMUST00000073663.3
|
Olfr1408
|
olfactory receptor 1408 |
| chr14_-_70761507 | 0.01 |
ENSMUST00000022692.5
|
Sftpc
|
surfactant associated protein C |
| chr18_+_37433852 | 0.01 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr19_-_10655391 | 0.01 |
ENSMUST00000025647.7
|
Pga5
|
pepsinogen 5, group I |
| chr14_-_50322136 | 0.01 |
ENSMUST00000072370.3
|
Olfr726
|
olfactory receptor 726 |
| chr7_+_123061497 | 0.01 |
ENSMUST00000033023.10
|
Aqp8
|
aquaporin 8 |
| chr7_-_103261741 | 0.01 |
ENSMUST00000052152.3
|
Olfr620
|
olfactory receptor 620 |
| chr12_+_78243846 | 0.01 |
ENSMUST00000188791.2
|
Gm6657
|
predicted gene 6657 |
| chr13_-_103042294 | 0.01 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_-_111608339 | 0.01 |
ENSMUST00000039887.4
|
Pof1b
|
premature ovarian failure 1B |
| chr13_+_27241551 | 0.01 |
ENSMUST00000110369.10
ENSMUST00000224228.2 ENSMUST00000018061.7 |
Prl
|
prolactin |
| chr13_-_22403990 | 0.01 |
ENSMUST00000057516.2
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr18_+_76944006 | 0.01 |
ENSMUST00000166956.2
|
Skor2
|
SKI family transcriptional corepressor 2 |
| chr2_+_74535242 | 0.01 |
ENSMUST00000019749.4
|
Hoxd8
|
homeobox D8 |
| chr7_+_106673037 | 0.01 |
ENSMUST00000054629.7
|
Olfr714
|
olfactory receptor 714 |
| chr17_+_38378064 | 0.01 |
ENSMUST00000087129.3
|
Olfr130
|
olfactory receptor 130 |
| chr10_+_101517556 | 0.01 |
ENSMUST00000156751.8
|
Mgat4c
|
MGAT4 family, member C |
| chr6_+_87405968 | 0.01 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr2_+_74534959 | 0.01 |
ENSMUST00000151380.2
|
Hoxd8
|
homeobox D8 |
| chr3_-_59170245 | 0.01 |
ENSMUST00000050360.14
ENSMUST00000199609.2 |
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr1_-_28819331 | 0.01 |
ENSMUST00000059937.5
|
Gm597
|
predicted gene 597 |
| chr8_-_84771610 | 0.01 |
ENSMUST00000061923.5
|
Rln3
|
relaxin 3 |
| chr8_+_57774010 | 0.01 |
ENSMUST00000040104.5
|
Hand2
|
heart and neural crest derivatives expressed 2 |
| chr16_-_34083315 | 0.00 |
ENSMUST00000114953.8
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr19_+_58658779 | 0.00 |
ENSMUST00000057270.9
|
Pnlip
|
pancreatic lipase |
| chr15_-_101726600 | 0.00 |
ENSMUST00000023712.8
|
Krt2
|
keratin 2 |
| chr1_-_14380418 | 0.00 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_-_95697441 | 0.00 |
ENSMUST00000119760.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr9_-_40039335 | 0.00 |
ENSMUST00000060345.6
|
Olfr985
|
olfactory receptor 985 |
| chr19_+_12405268 | 0.00 |
ENSMUST00000168148.2
|
Pfpl
|
pore forming protein-like |
| chr19_+_58658838 | 0.00 |
ENSMUST00000238108.2
|
Pnlip
|
pancreatic lipase |
| chr10_+_116881246 | 0.00 |
ENSMUST00000073834.5
|
Lrrc10
|
leucine rich repeat containing 10 |
| chr9_+_46139878 | 0.00 |
ENSMUST00000034588.9
ENSMUST00000132155.2 |
Apoa1
|
apolipoprotein A-I |
| chr3_-_30194559 | 0.00 |
ENSMUST00000108271.10
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr19_-_11848669 | 0.00 |
ENSMUST00000087857.3
|
Olfr1419
|
olfactory receptor 1419 |
| chr10_-_23226684 | 0.00 |
ENSMUST00000220299.2
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.1 | 0.9 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0032377 | dolichol metabolic process(GO:0019348) regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |