Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Gbx2
Z-value: 0.62
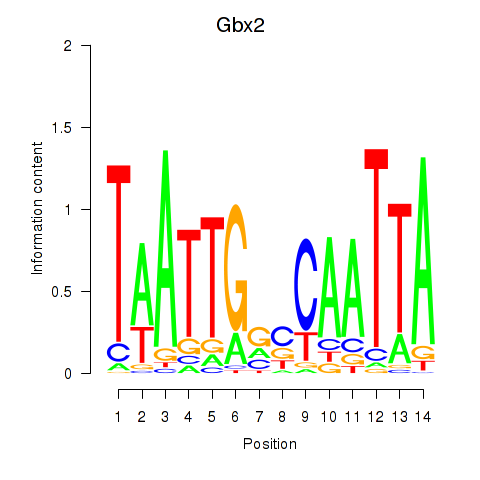
Transcription factors associated with Gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gbx2
|
ENSMUSG00000034486.9 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gbx2 | mm39_v1_chr1_-_89858901_89858920 | -0.93 | 2.3e-02 | Click! |
Activity profile of Gbx2 motif
Sorted Z-values of Gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23947641 | 1.09 |
ENSMUST00000055770.4
|
H1f1
|
H1.1 linker histone, cluster member |
| chr2_+_87609827 | 0.75 |
ENSMUST00000105210.3
|
Olfr152
|
olfactory receptor 152 |
| chr19_-_12313274 | 0.69 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1 |
| chrM_+_14138 | 0.53 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.53 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr17_-_37974666 | 0.45 |
ENSMUST00000215414.2
ENSMUST00000213638.3 |
Olfr117
|
olfactory receptor 117 |
| chrX_+_137815171 | 0.36 |
ENSMUST00000064937.14
ENSMUST00000113052.8 |
Nrk
|
Nik related kinase |
| chr4_-_139695337 | 0.31 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr11_+_97206542 | 0.31 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr2_-_85966272 | 0.31 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr2_-_87838612 | 0.29 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr1_-_156301821 | 0.24 |
ENSMUST00000188027.2
ENSMUST00000187507.7 ENSMUST00000189661.7 |
Soat1
|
sterol O-acyltransferase 1 |
| chr1_-_155293141 | 0.24 |
ENSMUST00000111775.8
ENSMUST00000111774.2 |
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr11_-_103158190 | 0.23 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr16_-_97883632 | 0.23 |
ENSMUST00000231268.2
ENSMUST00000231741.2 |
A630089N07Rik
|
RIKEN cDNA A630089N07 gene |
| chr1_+_92533504 | 0.22 |
ENSMUST00000217316.2
ENSMUST00000216553.2 |
Olfr1410
|
olfactory receptor 1410 |
| chr13_+_49697919 | 0.22 |
ENSMUST00000177948.2
ENSMUST00000021820.14 |
Aspn
|
asporin |
| chr11_-_58521327 | 0.21 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr8_+_72837021 | 0.21 |
ENSMUST00000213940.2
|
Olfr373
|
olfactory receptor 373 |
| chr5_+_113873864 | 0.21 |
ENSMUST00000065698.7
|
Ficd
|
FIC domain containing |
| chr2_+_84818538 | 0.20 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr7_-_140402037 | 0.20 |
ENSMUST00000106052.2
ENSMUST00000080651.13 |
Zfp941
|
zinc finger protein 941 |
| chr10_-_128918779 | 0.19 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr2_-_111324108 | 0.17 |
ENSMUST00000208881.2
ENSMUST00000208695.2 ENSMUST00000217611.2 |
Olfr1290
|
olfactory receptor 1290 |
| chr4_+_118818775 | 0.16 |
ENSMUST00000058651.5
|
Lao1
|
L-amino acid oxidase 1 |
| chr10_-_95337783 | 0.14 |
ENSMUST00000075829.3
ENSMUST00000217777.2 ENSMUST00000218893.2 |
Mrpl42
|
mitochondrial ribosomal protein L42 |
| chr7_-_12829100 | 0.13 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr2_+_152596075 | 0.13 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr18_+_57487908 | 0.13 |
ENSMUST00000238069.2
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr3_+_84573499 | 0.12 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr9_+_108708939 | 0.11 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr11_-_79394904 | 0.09 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr16_-_26190578 | 0.09 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr11_-_73707875 | 0.09 |
ENSMUST00000118463.4
ENSMUST00000144724.3 |
Olfr392
|
olfactory receptor 392 |
| chr7_-_105230807 | 0.09 |
ENSMUST00000191011.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr2_+_70392351 | 0.08 |
ENSMUST00000094934.11
|
Gad1
|
glutamate decarboxylase 1 |
| chr7_-_105230698 | 0.07 |
ENSMUST00000189378.7
|
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr3_-_96359622 | 0.07 |
ENSMUST00000093126.11
ENSMUST00000098841.4 |
BC107364
|
cDNA sequence BC107364 |
| chr14_+_45225641 | 0.07 |
ENSMUST00000046891.6
|
Ptger2
|
prostaglandin E receptor 2 (subtype EP2) |
| chr5_-_5315968 | 0.05 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr7_-_105230395 | 0.05 |
ENSMUST00000188726.2
ENSMUST00000188440.7 |
Apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 |
| chr1_-_52991993 | 0.05 |
ENSMUST00000050567.11
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr4_+_32657105 | 0.05 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chrX_+_105964224 | 0.04 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr2_-_73410632 | 0.04 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr12_-_98225676 | 0.03 |
ENSMUST00000021390.9
|
Galc
|
galactosylceramidase |
| chr4_-_26346882 | 0.03 |
ENSMUST00000041374.8
ENSMUST00000153813.2 |
Manea
|
mannosidase, endo-alpha |
| chr14_-_66451001 | 0.02 |
ENSMUST00000178730.8
|
Ptk2b
|
PTK2 protein tyrosine kinase 2 beta |
| chr3_-_86827664 | 0.02 |
ENSMUST00000194452.2
ENSMUST00000191752.6 |
Dclk2
|
doublecortin-like kinase 2 |
| chr12_+_79344119 | 0.02 |
ENSMUST00000079533.12
|
Rad51b
|
RAD51 paralog B |
| chr4_+_110254858 | 0.02 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr2_+_70392491 | 0.01 |
ENSMUST00000148210.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr10_+_129610507 | 0.01 |
ENSMUST00000203598.3
|
Olfr809
|
olfactory receptor 809 |
| chr10_-_5144699 | 0.01 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr7_+_28510310 | 0.01 |
ENSMUST00000038572.15
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr4_+_110254907 | 0.01 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr8_-_65471175 | 0.01 |
ENSMUST00000078409.5
ENSMUST00000048565.9 |
Trim61
Trim60
|
tripartite motif-containing 61 tripartite motif-containing 60 |
| chr11_-_46597885 | 0.01 |
ENSMUST00000055102.13
ENSMUST00000125008.2 |
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr1_-_74932266 | 0.01 |
ENSMUST00000006721.3
|
Cryba2
|
crystallin, beta A2 |
| chr9_-_40875414 | 0.01 |
ENSMUST00000034521.8
|
Jhy
|
junctional cadherin complex regulator |
| chr16_-_13548833 | 0.01 |
ENSMUST00000023364.7
|
Pla2g10
|
phospholipase A2, group X |
| chr1_-_161807205 | 0.00 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr9_+_37656402 | 0.00 |
ENSMUST00000216982.2
|
Olfr874
|
olfactory receptor 874 |
| chr14_-_48904958 | 0.00 |
ENSMUST00000144465.8
ENSMUST00000133479.8 ENSMUST00000119070.8 ENSMUST00000226501.2 |
Otx2
|
orthodenticle homeobox 2 |
| chr2_-_85632888 | 0.00 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr3_-_86827640 | 0.00 |
ENSMUST00000195561.6
|
Dclk2
|
doublecortin-like kinase 2 |
| chr19_+_38121214 | 0.00 |
ENSMUST00000112329.3
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr18_-_78640066 | 0.00 |
ENSMUST00000235389.2
ENSMUST00000237674.2 |
Slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr19_+_38121248 | 0.00 |
ENSMUST00000025956.13
|
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr5_+_7354130 | 0.00 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.2 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0050760 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.2 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |