Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
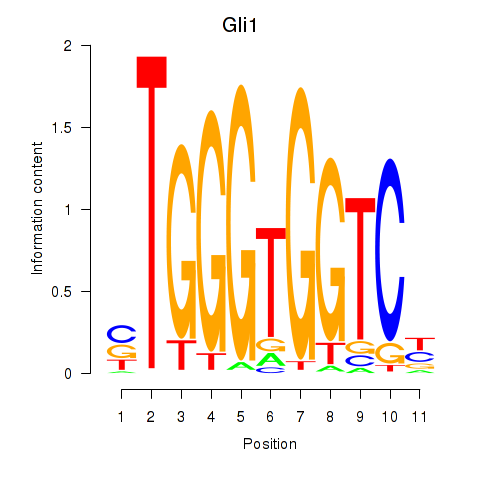
Results for Gli1
Z-value: 1.28
Transcription factors associated with Gli1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gli1
|
ENSMUSG00000025407.8 | GLI-Kruppel family member GLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gli1 | mm39_v1_chr10_-_127177729_127177843 | -0.91 | 3.4e-02 | Click! |
Activity profile of Gli1 motif
Sorted Z-values of Gli1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34218301 | 0.94 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr11_-_96714813 | 0.90 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr6_-_124410452 | 0.79 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr10_-_34003933 | 0.76 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr12_+_84363603 | 0.68 |
ENSMUST00000045931.12
|
Zfp410
|
zinc finger protein 410 |
| chr7_+_24069680 | 0.68 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chrX_-_166165743 | 0.65 |
ENSMUST00000026839.5
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr4_-_154983533 | 0.61 |
ENSMUST00000030935.10
ENSMUST00000132281.2 |
Prxl2b
|
peroxiredoxin like 2B |
| chr17_-_90217868 | 0.58 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr5_-_115438971 | 0.58 |
ENSMUST00000112090.2
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr9_+_106083988 | 0.56 |
ENSMUST00000188650.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr15_-_103218876 | 0.56 |
ENSMUST00000079824.6
|
Gpr84
|
G protein-coupled receptor 84 |
| chr11_-_76289888 | 0.53 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr17_+_34406762 | 0.53 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chrX_-_141173330 | 0.53 |
ENSMUST00000112907.8
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr6_-_52195663 | 0.52 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr14_+_55798362 | 0.49 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr17_+_34406523 | 0.49 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_+_30061469 | 0.49 |
ENSMUST00000015481.6
|
Endog
|
endonuclease G |
| chr18_-_35795233 | 0.48 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr4_+_106418224 | 0.48 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr19_-_46321218 | 0.46 |
ENSMUST00000238062.2
|
Cuedc2
|
CUE domain containing 2 |
| chr11_+_115310885 | 0.44 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr7_-_110673269 | 0.44 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr13_+_81931196 | 0.43 |
ENSMUST00000022009.10
ENSMUST00000223793.2 |
Cetn3
|
centrin 3 |
| chrX_+_158086253 | 0.43 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr14_+_105496008 | 0.42 |
ENSMUST00000181969.8
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr5_-_110927803 | 0.42 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr17_-_85097945 | 0.41 |
ENSMUST00000112308.9
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr14_+_55798517 | 0.40 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_105496147 | 0.38 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr3_-_94566107 | 0.38 |
ENSMUST00000196655.5
ENSMUST00000200407.2 ENSMUST00000006123.11 ENSMUST00000196733.5 |
Tuft1
|
tuftelin 1 |
| chr7_-_139696308 | 0.37 |
ENSMUST00000026538.13
|
Echs1
|
enoyl Coenzyme A hydratase, short chain, 1, mitochondrial |
| chr6_-_67014348 | 0.37 |
ENSMUST00000204369.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr6_+_95094721 | 0.37 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr12_-_69243871 | 0.37 |
ENSMUST00000223192.2
|
Gm49383
|
predicted gene, 49383 |
| chr10_+_79613083 | 0.37 |
ENSMUST00000020575.5
|
Fstl3
|
follistatin-like 3 |
| chr18_-_35795175 | 0.35 |
ENSMUST00000236574.2
ENSMUST00000236971.2 |
Spata24
|
spermatogenesis associated 24 |
| chr5_-_123038329 | 0.35 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr8_-_106692668 | 0.34 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chrX_-_141173487 | 0.33 |
ENSMUST00000112904.8
ENSMUST00000112903.8 ENSMUST00000033634.5 |
Acsl4
|
acyl-CoA synthetase long-chain family member 4 |
| chr11_+_115310963 | 0.32 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr17_-_23783063 | 0.31 |
ENSMUST00000095606.5
|
Zfp213
|
zinc finger protein 213 |
| chr6_+_28423539 | 0.31 |
ENSMUST00000020717.12
ENSMUST00000169841.2 |
Arf5
|
ADP-ribosylation factor 5 |
| chr5_-_110987441 | 0.31 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr13_+_81931642 | 0.30 |
ENSMUST00000224574.2
|
Cetn3
|
centrin 3 |
| chr12_+_17778198 | 0.30 |
ENSMUST00000222944.2
|
Hpcal1
|
hippocalcin-like 1 |
| chr3_+_95836558 | 0.30 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_-_139857424 | 0.30 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr4_-_117539431 | 0.30 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr8_-_41494890 | 0.30 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr7_-_79989900 | 0.30 |
ENSMUST00000133728.8
|
Unc45a
|
unc-45 myosin chaperone A |
| chr5_-_110987604 | 0.29 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr11_+_23256883 | 0.29 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr7_+_101034604 | 0.28 |
ENSMUST00000130016.8
ENSMUST00000134143.2 |
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr9_+_32607301 | 0.27 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr13_-_63579497 | 0.27 |
ENSMUST00000160931.2
ENSMUST00000099444.10 ENSMUST00000220684.2 ENSMUST00000161977.8 ENSMUST00000163091.8 |
Fancc
|
Fanconi anemia, complementation group C |
| chr14_+_55797934 | 0.26 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_-_126074222 | 0.26 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
| chr11_+_23256909 | 0.26 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_+_4085164 | 0.25 |
ENSMUST00000003677.11
ENSMUST00000145705.8 |
Rnf215
|
ring finger protein 215 |
| chr7_-_126073994 | 0.24 |
ENSMUST00000205733.2
ENSMUST00000205889.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr11_+_78136569 | 0.23 |
ENSMUST00000002133.9
|
Sdf2
|
stromal cell derived factor 2 |
| chr19_-_21418215 | 0.23 |
ENSMUST00000237823.2
|
Gda
|
guanine deaminase |
| chr8_-_96161414 | 0.22 |
ENSMUST00000211908.2
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr7_+_139695707 | 0.22 |
ENSMUST00000211757.2
|
Paox
|
polyamine oxidase (exo-N4-amino) |
| chr2_-_152672535 | 0.22 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr6_-_148846247 | 0.21 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr6_-_67014191 | 0.21 |
ENSMUST00000204282.2
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chrX_+_154045439 | 0.20 |
ENSMUST00000026324.10
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr9_-_106762818 | 0.20 |
ENSMUST00000185707.2
|
Rbm15b
|
RNA binding motif protein 15B |
| chr2_+_26220403 | 0.20 |
ENSMUST00000114134.9
ENSMUST00000127453.2 |
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr9_-_96774719 | 0.20 |
ENSMUST00000154146.8
|
Pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr12_+_84363876 | 0.20 |
ENSMUST00000222258.2
ENSMUST00000222832.2 ENSMUST00000220931.2 |
Zfp410
|
zinc finger protein 410 |
| chr13_+_54722833 | 0.20 |
ENSMUST00000156024.2
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr4_-_45320579 | 0.19 |
ENSMUST00000030003.10
|
Exosc3
|
exosome component 3 |
| chr5_-_134485081 | 0.19 |
ENSMUST00000111244.5
|
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr1_-_84817000 | 0.19 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_+_29780122 | 0.19 |
ENSMUST00000113762.8
ENSMUST00000113765.8 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr2_+_181139016 | 0.19 |
ENSMUST00000108799.10
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr2_+_119181703 | 0.19 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr9_+_103940879 | 0.19 |
ENSMUST00000047799.13
ENSMUST00000189998.3 |
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr11_-_34724458 | 0.19 |
ENSMUST00000093191.3
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr5_-_123127346 | 0.19 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr10_-_128727542 | 0.19 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr1_-_106641940 | 0.19 |
ENSMUST00000112751.2
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr9_-_108183162 | 0.18 |
ENSMUST00000044725.9
|
Tcta
|
T cell leukemia translocation altered gene |
| chrX_+_135950334 | 0.18 |
ENSMUST00000047852.8
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr10_+_79722081 | 0.18 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr2_+_181138958 | 0.18 |
ENSMUST00000149163.8
ENSMUST00000000844.15 ENSMUST00000184849.8 ENSMUST00000108800.8 ENSMUST00000069712.9 |
Tpd52l2
|
tumor protein D52-like 2 |
| chr4_-_133601990 | 0.18 |
ENSMUST00000168974.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_+_3239868 | 0.17 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_+_24310171 | 0.17 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr9_-_54569128 | 0.17 |
ENSMUST00000034822.12
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr3_+_95836637 | 0.16 |
ENSMUST00000171368.8
ENSMUST00000168106.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_12140263 | 0.16 |
ENSMUST00000050069.9
ENSMUST00000069128.8 |
Rbm12b1
|
RNA binding motif protein 12 B1 |
| chr2_-_92264374 | 0.16 |
ENSMUST00000111278.2
ENSMUST00000090559.12 |
Cry2
|
cryptochrome 2 (photolyase-like) |
| chr3_+_68401536 | 0.16 |
ENSMUST00000182719.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr13_-_53531391 | 0.16 |
ENSMUST00000021920.8
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr8_+_77628916 | 0.16 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr2_+_84867554 | 0.16 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr7_+_79836581 | 0.15 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chrX_+_139857640 | 0.15 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr13_+_6598185 | 0.15 |
ENSMUST00000021611.10
ENSMUST00000222485.2 |
Pitrm1
|
pitrilysin metallepetidase 1 |
| chr12_+_44375747 | 0.15 |
ENSMUST00000020939.16
ENSMUST00000220126.2 |
Nrcam
|
neuronal cell adhesion molecule |
| chrX_+_139857688 | 0.15 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr7_+_140796537 | 0.14 |
ENSMUST00000141804.2
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr10_+_80085275 | 0.14 |
ENSMUST00000020361.7
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr6_-_113508536 | 0.13 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr14_+_19801333 | 0.13 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2 |
| chr12_+_77285770 | 0.13 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr14_-_40730180 | 0.13 |
ENSMUST00000136661.8
|
Prxl2a
|
peroxiredoxin like 2A |
| chr2_+_129642371 | 0.12 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr15_-_51855073 | 0.12 |
ENSMUST00000022927.11
|
Rad21
|
RAD21 cohesin complex component |
| chr2_+_76200299 | 0.12 |
ENSMUST00000046389.5
|
Rbm45
|
RNA binding motif protein 45 |
| chr7_+_25016492 | 0.12 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr2_-_152673032 | 0.12 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr6_+_113508636 | 0.12 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr8_-_41469332 | 0.12 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr2_-_121287174 | 0.12 |
ENSMUST00000110613.9
ENSMUST00000056312.10 |
Serinc4
|
serine incorporator 4 |
| chr2_+_20742115 | 0.11 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr2_-_91762033 | 0.11 |
ENSMUST00000111309.8
ENSMUST00000090602.6 |
Mdk
|
midkine |
| chr9_+_113570506 | 0.11 |
ENSMUST00000213663.2
|
Clasp2
|
CLIP associating protein 2 |
| chr2_+_29779750 | 0.11 |
ENSMUST00000113763.8
ENSMUST00000113757.8 ENSMUST00000113756.8 ENSMUST00000133233.8 ENSMUST00000113759.9 ENSMUST00000113755.8 ENSMUST00000137558.8 ENSMUST00000046571.14 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr2_+_29780073 | 0.11 |
ENSMUST00000028128.13
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr15_+_99290763 | 0.11 |
ENSMUST00000023749.15
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_-_6334239 | 0.11 |
ENSMUST00000127658.2
ENSMUST00000062765.14 |
Zfp583
|
zinc finger protein 583 |
| chr16_+_10229786 | 0.11 |
ENSMUST00000023146.5
|
Nubp1
|
nucleotide binding protein 1 |
| chr12_+_44375665 | 0.11 |
ENSMUST00000110748.4
|
Nrcam
|
neuronal cell adhesion molecule |
| chr7_+_27198740 | 0.10 |
ENSMUST00000098644.9
ENSMUST00000108355.2 ENSMUST00000238936.2 |
Prx
|
periaxin |
| chr2_-_91762119 | 0.10 |
ENSMUST00000069423.13
|
Mdk
|
midkine |
| chr7_+_28682253 | 0.10 |
ENSMUST00000085835.8
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr6_-_52222776 | 0.10 |
ENSMUST00000048026.10
|
Hoxa11
|
homeobox A11 |
| chr5_-_115439016 | 0.09 |
ENSMUST00000009157.4
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr4_-_43454561 | 0.09 |
ENSMUST00000107926.8
ENSMUST00000107925.8 |
Cd72
|
CD72 antigen |
| chr17_-_34247016 | 0.09 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr12_-_113945961 | 0.08 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr9_+_110134241 | 0.08 |
ENSMUST00000199592.5
ENSMUST00000068071.12 |
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr2_+_29780532 | 0.08 |
ENSMUST00000113764.4
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr10_+_69369590 | 0.08 |
ENSMUST00000182884.8
|
Ank3
|
ankyrin 3, epithelial |
| chr5_-_32903615 | 0.07 |
ENSMUST00000120591.8
|
Pisd
|
phosphatidylserine decarboxylase |
| chr2_+_130748380 | 0.07 |
ENSMUST00000028781.9
|
Atrn
|
attractin |
| chr11_+_4085233 | 0.07 |
ENSMUST00000124670.2
|
Rnf215
|
ring finger protein 215 |
| chr9_-_108183356 | 0.07 |
ENSMUST00000192886.6
|
Tcta
|
T cell leukemia translocation altered gene |
| chr2_+_84867783 | 0.07 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr17_+_37356872 | 0.06 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr12_-_70158348 | 0.06 |
ENSMUST00000220689.2
|
Nin
|
ninein |
| chr3_+_115801106 | 0.06 |
ENSMUST00000029575.12
ENSMUST00000106501.8 |
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr12_-_114419703 | 0.06 |
ENSMUST00000193397.2
|
Ighv6-7
|
immunoglobulin heavy variable V6-7 |
| chr4_+_118285275 | 0.05 |
ENSMUST00000006557.13
ENSMUST00000167636.8 ENSMUST00000102673.11 |
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 |
| chr13_-_114595475 | 0.05 |
ENSMUST00000022287.8
ENSMUST00000223640.3 |
Fst
|
follistatin |
| chr7_-_19043525 | 0.05 |
ENSMUST00000208446.2
|
Fosb
|
FBJ osteosarcoma oncogene B |
| chr7_+_140796559 | 0.05 |
ENSMUST00000148975.3
|
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr10_-_63257568 | 0.05 |
ENSMUST00000054760.6
|
Gm7075
|
predicted gene 7075 |
| chr9_+_117888124 | 0.05 |
ENSMUST00000123690.2
|
Azi2
|
5-azacytidine induced gene 2 |
| chr10_+_69369632 | 0.04 |
ENSMUST00000182155.8
ENSMUST00000183169.8 ENSMUST00000183148.8 |
Ank3
|
ankyrin 3, epithelial |
| chr10_-_43934774 | 0.04 |
ENSMUST00000239010.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr17_+_34457868 | 0.04 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr8_-_96161466 | 0.04 |
ENSMUST00000213086.2
ENSMUST00000034249.8 |
Cfap20
|
cilia and flagella associated protein 20 |
| chr5_-_146900548 | 0.04 |
ENSMUST00000125217.2
ENSMUST00000110564.8 ENSMUST00000066675.10 ENSMUST00000016654.9 ENSMUST00000110566.2 ENSMUST00000140526.2 |
Mtif3
|
mitochondrial translational initiation factor 3 |
| chr11_-_45845992 | 0.04 |
ENSMUST00000109254.2
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr1_+_125488747 | 0.04 |
ENSMUST00000027580.11
|
Slc35f5
|
solute carrier family 35, member F5 |
| chr2_+_118865262 | 0.04 |
ENSMUST00000028796.2
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr4_-_40948196 | 0.03 |
ENSMUST00000030125.5
ENSMUST00000108089.8 ENSMUST00000191273.7 |
Bag1
|
BCL2-associated athanogene 1 |
| chr4_+_62398262 | 0.03 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr4_+_116953218 | 0.03 |
ENSMUST00000030443.12
|
Ptch2
|
patched 2 |
| chr16_-_19703014 | 0.03 |
ENSMUST00000100083.5
ENSMUST00000231564.2 |
A930003A15Rik
|
RIKEN cDNA A930003A15 gene |
| chrX_-_58612709 | 0.03 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr11_-_115310743 | 0.03 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr2_-_87734018 | 0.02 |
ENSMUST00000099844.2
|
Olfr1154
|
olfactory receptor 1154 |
| chr8_+_46111703 | 0.02 |
ENSMUST00000134675.8
ENSMUST00000139869.8 ENSMUST00000126067.8 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_-_66915756 | 0.02 |
ENSMUST00000207715.2
|
Mef2a
|
myocyte enhancer factor 2A |
| chr11_+_118324652 | 0.02 |
ENSMUST00000106286.3
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr8_+_55407872 | 0.02 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr10_+_69370038 | 0.02 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr6_+_107506678 | 0.02 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr11_+_9141934 | 0.02 |
ENSMUST00000042740.13
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr19_-_42740898 | 0.02 |
ENSMUST00000237747.2
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr2_+_106523532 | 0.02 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr13_-_114595122 | 0.02 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chrX_-_111315519 | 0.02 |
ENSMUST00000124335.8
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr19_+_13745671 | 0.02 |
ENSMUST00000061669.3
|
Olfr1495
|
olfactory receptor 1495 |
| chr9_-_54568950 | 0.02 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr19_+_4771089 | 0.02 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr11_-_42072990 | 0.01 |
ENSMUST00000205546.2
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 |
| chr16_+_92295009 | 0.01 |
ENSMUST00000023670.4
|
Clic6
|
chloride intracellular channel 6 |
| chr14_-_52648384 | 0.01 |
ENSMUST00000079459.4
|
Olfr1510
|
olfactory receptor 1510 |
| chr6_-_41613322 | 0.01 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr14_-_54647647 | 0.01 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr2_-_35994819 | 0.01 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr12_-_113876103 | 0.01 |
ENSMUST00000193133.3
ENSMUST00000103462.4 |
Ighv7-2
|
immunoglobulin heavy variable 7-2 |
| chr11_-_94595325 | 0.01 |
ENSMUST00000069852.2
|
Gm11541
|
predicted gene 11541 |
| chr4_-_58553311 | 0.01 |
ENSMUST00000107571.8
ENSMUST00000055018.11 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr8_+_46111778 | 0.01 |
ENSMUST00000143820.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_+_37356854 | 0.01 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr12_-_113823290 | 0.01 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr7_-_99340830 | 0.01 |
ENSMUST00000208713.2
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr1_-_25267894 | 0.01 |
ENSMUST00000126626.8
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr1_+_129201081 | 0.01 |
ENSMUST00000073527.13
ENSMUST00000040311.14 |
Thsd7b
|
thrombospondin, type I, domain containing 7B |
| chr11_-_41891359 | 0.01 |
ENSMUST00000070735.10
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gli1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 0.9 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 | 0.9 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.2 | 0.7 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.5 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.5 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.2 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) |
| 0.1 | 0.7 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.1 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.6 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.1 | 0.2 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.3 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.3 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.1 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.0 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.4 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.9 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |