Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
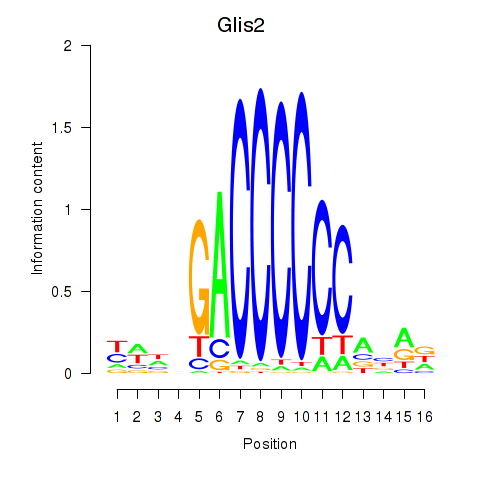
Results for Glis2
Z-value: 0.92
Transcription factors associated with Glis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis2
|
ENSMUSG00000014303.14 | GLIS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis2 | mm39_v1_chr16_+_4412546_4412599 | -0.84 | 7.2e-02 | Click! |
Activity profile of Glis2 motif
Sorted Z-values of Glis2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_126303887 | 0.76 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr11_+_87684548 | 0.70 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr7_+_127345909 | 0.52 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr17_-_65901946 | 0.51 |
ENSMUST00000232686.2
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr17_+_26882171 | 0.51 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr10_+_79650496 | 0.50 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr5_-_137531463 | 0.46 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_-_93729562 | 0.45 |
ENSMUST00000112890.3
|
Dtymk
|
deoxythymidylate kinase |
| chr4_-_15945359 | 0.43 |
ENSMUST00000029877.9
|
Decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr11_+_4823951 | 0.43 |
ENSMUST00000038570.9
|
Nipsnap1
|
nipsnap homolog 1 |
| chr7_+_141056305 | 0.42 |
ENSMUST00000117634.2
|
Tspan4
|
tetraspanin 4 |
| chr6_-_124410452 | 0.42 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr16_+_32698470 | 0.40 |
ENSMUST00000232272.2
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr5_-_137531413 | 0.39 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_-_118239146 | 0.38 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr2_+_90507552 | 0.37 |
ENSMUST00000057481.7
|
Nup160
|
nucleoporin 160 |
| chr12_-_31684588 | 0.37 |
ENSMUST00000020979.9
ENSMUST00000177962.9 |
Bcap29
|
B cell receptor associated protein 29 |
| chr5_-_124490296 | 0.36 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr11_+_98932586 | 0.35 |
ENSMUST00000177092.8
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr17_-_26014613 | 0.35 |
ENSMUST00000235889.2
|
Gm50367
|
predicted gene, 50367 |
| chr12_-_80690573 | 0.34 |
ENSMUST00000166931.2
ENSMUST00000218364.2 |
Erh
|
ERH mRNA splicing and mitosis factor |
| chr6_-_29212295 | 0.34 |
ENSMUST00000078155.12
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr17_+_7246365 | 0.32 |
ENSMUST00000232245.2
|
Rnaset2b
|
ribonuclease T2B |
| chr14_-_31362835 | 0.31 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr7_+_27290969 | 0.30 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr10_+_127157784 | 0.30 |
ENSMUST00000219511.2
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr1_-_131204651 | 0.28 |
ENSMUST00000161764.8
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr14_+_55120875 | 0.27 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr6_-_29212239 | 0.27 |
ENSMUST00000160878.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr3_+_94882142 | 0.27 |
ENSMUST00000167008.8
ENSMUST00000107251.9 |
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr17_+_7246289 | 0.26 |
ENSMUST00000179728.2
|
Rnaset2b
|
ribonuclease T2B |
| chr5_-_134643805 | 0.26 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr9_+_75213570 | 0.25 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr5_+_115417725 | 0.25 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chrX_-_166165743 | 0.24 |
ENSMUST00000026839.5
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr9_+_21634779 | 0.24 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr9_+_21634918 | 0.24 |
ENSMUST00000213114.2
|
Ldlr
|
low density lipoprotein receptor |
| chr7_+_27291126 | 0.24 |
ENSMUST00000167435.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr12_+_73170483 | 0.24 |
ENSMUST00000187549.7
ENSMUST00000021523.7 |
Mnat1
|
menage a trois 1 |
| chr17_-_8366536 | 0.24 |
ENSMUST00000231927.2
|
Rnaset2a
|
ribonuclease T2A |
| chrX_-_106446928 | 0.23 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr3_+_127584251 | 0.23 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr11_-_70560110 | 0.23 |
ENSMUST00000129434.2
ENSMUST00000018431.13 |
Spag7
|
sperm associated antigen 7 |
| chr10_+_127102193 | 0.22 |
ENSMUST00000026479.11
|
Dctn2
|
dynactin 2 |
| chr8_+_73488496 | 0.22 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr13_-_53083494 | 0.22 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr6_-_148732946 | 0.22 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr14_-_8536869 | 0.21 |
ENSMUST00000238865.2
ENSMUST00000159275.4 |
Gm11100
|
predicted gene 11100 |
| chr8_-_23747023 | 0.21 |
ENSMUST00000121783.7
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr17_-_31731222 | 0.21 |
ENSMUST00000236665.2
|
Wdr4
|
WD repeat domain 4 |
| chr1_+_135945798 | 0.20 |
ENSMUST00000117950.2
|
Tmem9
|
transmembrane protein 9 |
| chr10_+_13376745 | 0.20 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr1_-_84817000 | 0.20 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr19_+_45991907 | 0.20 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chr7_-_126398165 | 0.20 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr9_+_50768224 | 0.19 |
ENSMUST00000174628.8
ENSMUST00000034560.14 ENSMUST00000114437.9 ENSMUST00000175645.8 ENSMUST00000176349.8 ENSMUST00000176798.8 ENSMUST00000175640.8 |
Ppp2r1b
|
protein phosphatase 2, regulatory subunit A, beta |
| chr9_+_107852733 | 0.19 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr8_-_85413707 | 0.19 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr1_+_135945705 | 0.19 |
ENSMUST00000063719.15
|
Tmem9
|
transmembrane protein 9 |
| chr7_-_121700958 | 0.19 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr19_+_8713156 | 0.18 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr11_+_70416185 | 0.18 |
ENSMUST00000018430.7
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr9_+_25392843 | 0.18 |
ENSMUST00000040677.6
|
Eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr4_-_42661893 | 0.18 |
ENSMUST00000108006.4
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr10_+_93669680 | 0.18 |
ENSMUST00000216224.2
|
Usp44
|
ubiquitin specific peptidase 44 |
| chr16_+_17577464 | 0.18 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr2_-_152672535 | 0.17 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr6_+_136305516 | 0.17 |
ENSMUST00000204966.2
ENSMUST00000077886.7 |
Eif4a3l1
|
eukaryotic translation initiation factor 4A3 like 1 |
| chr17_+_34258411 | 0.17 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr6_-_48817675 | 0.17 |
ENSMUST00000203265.3
ENSMUST00000205159.3 |
Tmem176b
|
transmembrane protein 176B |
| chr14_-_31362909 | 0.17 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr5_+_125552878 | 0.16 |
ENSMUST00000031445.5
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr10_+_80971054 | 0.16 |
ENSMUST00000125261.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr7_-_141614758 | 0.16 |
ENSMUST00000211000.2
ENSMUST00000209725.2 ENSMUST00000084418.4 |
Mob2
|
MOB kinase activator 2 |
| chr10_+_36850532 | 0.15 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr14_+_31363004 | 0.15 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr7_+_144468837 | 0.15 |
ENSMUST00000128057.8
ENSMUST00000033388.12 ENSMUST00000141737.2 ENSMUST00000105895.2 |
LTO1
|
ABCE maturation factor |
| chr5_-_110987441 | 0.15 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr3_-_90416757 | 0.15 |
ENSMUST00000107343.8
ENSMUST00000001043.14 ENSMUST00000107344.8 ENSMUST00000076639.11 ENSMUST00000107346.8 ENSMUST00000146740.8 ENSMUST00000107342.2 ENSMUST00000049937.13 |
Chtop
|
chromatin target of PRMT1 |
| chr2_+_112092271 | 0.14 |
ENSMUST00000028553.4
|
Nop10
|
NOP10 ribonucleoprotein |
| chr11_-_102209767 | 0.14 |
ENSMUST00000174302.8
ENSMUST00000178839.8 ENSMUST00000006754.14 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr6_+_113581708 | 0.14 |
ENSMUST00000035725.7
|
Brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr8_-_72178340 | 0.14 |
ENSMUST00000153800.8
ENSMUST00000146100.8 |
Fcho1
|
FCH domain only 1 |
| chr10_+_126914755 | 0.13 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr4_+_118477994 | 0.12 |
ENSMUST00000030501.15
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr2_-_29142965 | 0.12 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr6_+_48818567 | 0.12 |
ENSMUST00000203639.3
|
Tmem176a
|
transmembrane protein 176A |
| chr10_-_128047658 | 0.12 |
ENSMUST00000061995.10
|
Spryd4
|
SPRY domain containing 4 |
| chr3_+_66888685 | 0.12 |
ENSMUST00000162036.8
|
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr4_-_128856213 | 0.12 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr7_+_25016492 | 0.11 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr16_+_17577493 | 0.11 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr14_+_54457978 | 0.11 |
ENSMUST00000103740.2
ENSMUST00000198398.5 |
Trac
|
T cell receptor alpha constant |
| chr5_-_110987604 | 0.11 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr1_-_93729650 | 0.11 |
ENSMUST00000027503.14
|
Dtymk
|
deoxythymidylate kinase |
| chr10_-_22607136 | 0.11 |
ENSMUST00000238910.2
ENSMUST00000127698.8 |
ENSMUSG00000118528.2
Tbpl1
|
novel protein TATA box binding protein-like 1 |
| chr19_+_7245591 | 0.11 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chr6_-_106777014 | 0.11 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr7_+_5060159 | 0.11 |
ENSMUST00000045543.8
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr7_-_19595221 | 0.10 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr2_+_119181703 | 0.10 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr6_-_51446850 | 0.10 |
ENSMUST00000069949.13
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr13_-_76205115 | 0.10 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chr2_-_152673032 | 0.10 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr8_-_120828211 | 0.10 |
ENSMUST00000034280.9
|
Zdhhc7
|
zinc finger, DHHC domain containing 7 |
| chr5_-_123662175 | 0.10 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr5_+_139238089 | 0.09 |
ENSMUST00000130326.8
|
Get4
|
golgi to ER traffic protein 4 |
| chr7_+_44221791 | 0.09 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase |
| chr19_+_6451667 | 0.09 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr11_-_102120917 | 0.09 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr6_-_51446752 | 0.09 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr3_+_121746862 | 0.08 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr6_-_54941673 | 0.08 |
ENSMUST00000203837.2
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chrX_+_35861851 | 0.08 |
ENSMUST00000073339.7
|
Pgrmc1
|
progesterone receptor membrane component 1 |
| chr14_+_61375893 | 0.08 |
ENSMUST00000089394.10
|
Sacs
|
sacsin |
| chr6_-_48818430 | 0.08 |
ENSMUST00000205147.3
|
Tmem176b
|
transmembrane protein 176B |
| chr4_-_58499398 | 0.07 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr12_-_81468705 | 0.07 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr2_+_153133326 | 0.07 |
ENSMUST00000028977.7
|
Kif3b
|
kinesin family member 3B |
| chr6_-_148732893 | 0.07 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr9_-_22218934 | 0.07 |
ENSMUST00000086278.7
ENSMUST00000215202.2 |
Zfp810
|
zinc finger protein 810 |
| chr7_-_92523396 | 0.07 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr5_-_38637474 | 0.07 |
ENSMUST00000143758.8
ENSMUST00000156272.8 |
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr15_+_41310815 | 0.06 |
ENSMUST00000090096.11
ENSMUST00000110297.10 |
Oxr1
|
oxidation resistance 1 |
| chr6_-_48818006 | 0.06 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B |
| chr5_+_24631829 | 0.06 |
ENSMUST00000155598.8
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr3_+_127584449 | 0.06 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr14_-_71003973 | 0.06 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr7_-_4909515 | 0.06 |
ENSMUST00000210663.2
|
Gm36210
|
predicted gene, 36210 |
| chr10_+_80641067 | 0.06 |
ENSMUST00000036016.6
|
Amh
|
anti-Mullerian hormone |
| chr6_+_120643323 | 0.06 |
ENSMUST00000112686.8
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr5_-_137531471 | 0.06 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr11_+_97340962 | 0.05 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr2_+_181243109 | 0.05 |
ENSMUST00000136481.8
ENSMUST00000002529.7 |
Prpf6
|
pre-mRNA splicing factor 6 |
| chr18_+_34910064 | 0.05 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr16_+_31918599 | 0.05 |
ENSMUST00000115168.9
|
Cep19
|
centrosomal protein 19 |
| chr8_-_23747057 | 0.05 |
ENSMUST00000051094.9
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chrX_+_149830166 | 0.05 |
ENSMUST00000026296.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr5_+_101912939 | 0.05 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr11_+_121593219 | 0.05 |
ENSMUST00000036742.14
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chrX_-_8072714 | 0.04 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr13_-_35211060 | 0.04 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr3_+_129974531 | 0.04 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr5_-_134343232 | 0.04 |
ENSMUST00000174867.8
|
Gtf2i
|
general transcription factor II I |
| chr19_+_45003304 | 0.04 |
ENSMUST00000039016.14
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr12_-_17061545 | 0.04 |
ENSMUST00000190691.2
|
2410004P03Rik
|
RIKEN cDNA 2410004P03 gene |
| chr1_+_34498836 | 0.04 |
ENSMUST00000027302.14
ENSMUST00000190122.2 |
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr12_-_113945961 | 0.04 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr12_-_114419703 | 0.04 |
ENSMUST00000193397.2
|
Ighv6-7
|
immunoglobulin heavy variable V6-7 |
| chr2_-_154411765 | 0.04 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chr4_-_118477960 | 0.03 |
ENSMUST00000071972.11
|
Cfap57
|
cilia and flagella associated protein 57 |
| chr4_-_117772163 | 0.03 |
ENSMUST00000036156.6
|
Ipo13
|
importin 13 |
| chr19_-_6947112 | 0.03 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr18_-_35841435 | 0.03 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr7_-_126014027 | 0.03 |
ENSMUST00000032968.7
ENSMUST00000206325.2 |
Cd19
|
CD19 antigen |
| chr8_+_3681127 | 0.03 |
ENSMUST00000159911.8
ENSMUST00000004745.9 |
Stxbp2
|
syntaxin binding protein 2 |
| chr5_+_136112817 | 0.03 |
ENSMUST00000100570.10
|
Rasa4
|
RAS p21 protein activator 4 |
| chr5_+_102916637 | 0.03 |
ENSMUST00000112852.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr5_-_143255713 | 0.03 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr7_+_45434755 | 0.03 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr12_-_54909568 | 0.03 |
ENSMUST00000078124.8
|
Cfl2
|
cofilin 2, muscle |
| chr8_-_88686188 | 0.02 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr11_+_101358990 | 0.02 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr2_+_164810139 | 0.02 |
ENSMUST00000202479.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr11_-_11987391 | 0.02 |
ENSMUST00000093321.12
|
Grb10
|
growth factor receptor bound protein 10 |
| chr16_-_58850641 | 0.02 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr12_-_17061504 | 0.02 |
ENSMUST00000189479.2
|
2410004P03Rik
|
RIKEN cDNA 2410004P03 gene |
| chr7_-_109322993 | 0.02 |
ENSMUST00000106735.9
ENSMUST00000033334.5 |
BC051019
|
cDNA sequence BC051019 |
| chr11_+_69909659 | 0.02 |
ENSMUST00000232002.2
ENSMUST00000134376.10 ENSMUST00000231221.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr15_-_78428865 | 0.02 |
ENSMUST00000053239.4
|
Sstr3
|
somatostatin receptor 3 |
| chr16_-_31918485 | 0.02 |
ENSMUST00000189013.8
ENSMUST00000155966.8 ENSMUST00000096109.11 ENSMUST00000232321.2 |
Pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr6_-_4747066 | 0.02 |
ENSMUST00000090686.11
ENSMUST00000133306.8 |
Sgce
|
sarcoglycan, epsilon |
| chr2_-_89361831 | 0.02 |
ENSMUST00000144885.3
|
Olfr1243
|
olfactory receptor 1243 |
| chr12_-_75224099 | 0.02 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr19_+_16416664 | 0.02 |
ENSMUST00000237350.2
|
Gna14
|
guanine nucleotide binding protein, alpha 14 |
| chr7_-_114235506 | 0.01 |
ENSMUST00000205714.2
ENSMUST00000206853.2 ENSMUST00000205933.2 ENSMUST00000206156.2 ENSMUST00000032907.9 ENSMUST00000032906.11 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr3_-_66888474 | 0.01 |
ENSMUST00000162439.8
ENSMUST00000162098.9 |
Shox2
|
short stature homeobox 2 |
| chr7_+_141755098 | 0.01 |
ENSMUST00000187512.2
ENSMUST00000084414.6 |
Krtap5-3
|
keratin associated protein 5-3 |
| chr3_+_87855973 | 0.01 |
ENSMUST00000005019.6
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr5_+_95021656 | 0.01 |
ENSMUST00000180076.2
|
Gm3183
|
predicted gene 3183 |
| chr13_+_22512081 | 0.01 |
ENSMUST00000091734.2
|
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr11_+_73819052 | 0.01 |
ENSMUST00000205791.2
|
Olfr396-ps1
|
olfactory receptor 396, pseudogene 1 |
| chr1_+_135764092 | 0.01 |
ENSMUST00000188028.7
ENSMUST00000178204.8 ENSMUST00000190451.7 ENSMUST00000189732.7 ENSMUST00000189355.7 |
Tnnt2
|
troponin T2, cardiac |
| chr6_-_90050000 | 0.01 |
ENSMUST00000071865.2
|
Vmn1r49
|
vomeronasal 1, receptor 49 |
| chr19_-_58442866 | 0.01 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr1_-_131204422 | 0.01 |
ENSMUST00000159195.2
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr4_+_155819257 | 0.01 |
ENSMUST00000147721.8
ENSMUST00000127188.3 |
Tmem240
|
transmembrane protein 240 |
| chr7_-_141744126 | 0.01 |
ENSMUST00000210290.2
ENSMUST00000084415.5 |
Gm10153
|
predicted gene 10153 |
| chr6_+_21215472 | 0.01 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr15_+_66763169 | 0.01 |
ENSMUST00000005255.9
|
Ccn4
|
cellular communication network factor 4 |
| chr7_-_23508625 | 0.01 |
ENSMUST00000166141.2
|
Vmn1r175
|
vomeronasal 1 receptor 175 |
| chr11_+_74174562 | 0.01 |
ENSMUST00000214048.3
ENSMUST00000143976.4 ENSMUST00000205790.2 ENSMUST00000206659.2 |
Olfr59
|
olfactory receptor 59 |
| chr11_+_76795346 | 0.01 |
ENSMUST00000072633.4
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chrX_+_8137372 | 0.01 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr3_+_100732768 | 0.01 |
ENSMUST00000054791.9
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr5_-_38316296 | 0.01 |
ENSMUST00000201415.4
|
Nsg1
|
neuron specific gene family member 1 |
| chr7_-_4999042 | 0.01 |
ENSMUST00000162502.2
|
Zfp579
|
zinc finger protein 579 |
| chr7_-_126303947 | 0.01 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr12_-_114117264 | 0.01 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr17_-_34337446 | 0.01 |
ENSMUST00000095347.13
|
Brd2
|
bromodomain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.2 | 0.6 | GO:0046072 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.2 | 0.5 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0090118 | regulation of phosphatidylcholine catabolic process(GO:0010899) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 0.8 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.5 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.5 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.2 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0061198 | maintenance of chromatin silencing(GO:0006344) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.5 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0090537 | CERF complex(GO:0090537) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0042165 | neurotransmitter binding(GO:0042165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |