Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Glis3
Z-value: 0.50
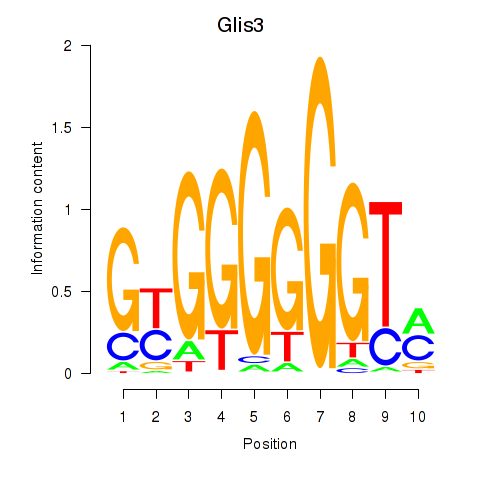
Transcription factors associated with Glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Glis3
|
ENSMUSG00000052942.14 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Glis3 | mm39_v1_chr19_-_28657477_28657523 | 0.53 | 3.6e-01 | Click! |
Activity profile of Glis3 motif
Sorted Z-values of Glis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_126380655 | 0.21 |
ENSMUST00000172352.8
ENSMUST00000094037.5 |
Tbx6
|
T-box 6 |
| chr2_+_32536594 | 0.20 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr8_+_23464860 | 0.17 |
ENSMUST00000110688.9
ENSMUST00000121802.9 |
Ank1
|
ankyrin 1, erythroid |
| chr2_+_156681991 | 0.17 |
ENSMUST00000073352.10
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr7_+_5023375 | 0.15 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865 |
| chr2_+_156681927 | 0.14 |
ENSMUST00000081335.13
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr11_+_98686321 | 0.14 |
ENSMUST00000037915.9
|
Msl1
|
male specific lethal 1 |
| chr13_-_40883893 | 0.13 |
ENSMUST00000021787.7
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr7_+_5023552 | 0.13 |
ENSMUST00000208728.2
ENSMUST00000085427.6 |
Ccdc106
Zfp865
|
coiled-coil domain containing 106 zinc finger protein 865 |
| chr4_-_114991478 | 0.12 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr4_+_137004793 | 0.10 |
ENSMUST00000045747.5
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr9_+_31191820 | 0.10 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr11_-_97635484 | 0.09 |
ENSMUST00000018691.9
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chr11_-_69692542 | 0.09 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr1_-_160862364 | 0.09 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr19_+_60878802 | 0.09 |
ENSMUST00000236876.2
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr8_+_106002772 | 0.09 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr1_+_166829001 | 0.09 |
ENSMUST00000126198.3
|
Fam78b
|
family with sequence similarity 78, member B |
| chr11_+_98818640 | 0.09 |
ENSMUST00000107474.8
|
Rara
|
retinoic acid receptor, alpha |
| chr1_+_166828982 | 0.09 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chrX_+_7439839 | 0.09 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr7_+_90091937 | 0.09 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr18_+_34910064 | 0.08 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr11_+_77873535 | 0.08 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr18_-_47501513 | 0.08 |
ENSMUST00000076043.13
ENSMUST00000135790.8 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_-_78228116 | 0.08 |
ENSMUST00000206268.2
ENSMUST00000039431.14 |
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr17_-_23964807 | 0.08 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr7_-_46445085 | 0.08 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr2_+_29780532 | 0.07 |
ENSMUST00000113764.4
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr13_+_38009981 | 0.07 |
ENSMUST00000110238.10
|
Rreb1
|
ras responsive element binding protein 1 |
| chr10_-_127124867 | 0.07 |
ENSMUST00000119078.8
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr7_-_15781838 | 0.07 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr10_-_13350106 | 0.07 |
ENSMUST00000105545.12
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr2_-_152673585 | 0.07 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr7_+_89779421 | 0.07 |
ENSMUST00000207225.2
ENSMUST00000207484.2 ENSMUST00000209068.2 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_-_152672535 | 0.07 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr8_+_109441276 | 0.07 |
ENSMUST00000043896.10
|
Zfhx3
|
zinc finger homeobox 3 |
| chr11_+_98686617 | 0.07 |
ENSMUST00000107485.8
ENSMUST00000107487.10 |
Msl1
|
male specific lethal 1 |
| chr5_-_91550853 | 0.07 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr19_-_47303184 | 0.07 |
ENSMUST00000237796.2
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr7_+_141276575 | 0.06 |
ENSMUST00000185406.8
|
Muc2
|
mucin 2 |
| chr7_+_89779493 | 0.06 |
ENSMUST00000208730.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr10_+_61010983 | 0.06 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr11_-_100510423 | 0.06 |
ENSMUST00000137688.9
ENSMUST00000239389.2 ENSMUST00000155152.9 ENSMUST00000154972.9 ENSMUST00000239479.2 ENSMUST00000132886.3 |
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr3_-_94489855 | 0.06 |
ENSMUST00000107283.8
|
Snx27
|
sorting nexin family member 27 |
| chr5_-_142803405 | 0.06 |
ENSMUST00000151477.8
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr9_+_21279299 | 0.06 |
ENSMUST00000214852.2
ENSMUST00000115414.3 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr4_+_115595610 | 0.06 |
ENSMUST00000106524.4
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr9_-_108183356 | 0.06 |
ENSMUST00000192886.6
|
Tcta
|
T cell leukemia translocation altered gene |
| chr7_+_79676095 | 0.06 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr5_+_137286535 | 0.05 |
ENSMUST00000024099.11
ENSMUST00000196208.5 ENSMUST00000085934.4 |
Ache
|
acetylcholinesterase |
| chrX_+_100298134 | 0.05 |
ENSMUST00000062000.6
|
Foxo4
|
forkhead box O4 |
| chr15_-_82783978 | 0.05 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20 |
| chr6_+_17463819 | 0.05 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr2_+_29780122 | 0.05 |
ENSMUST00000113762.8
ENSMUST00000113765.8 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr17_+_35191661 | 0.04 |
ENSMUST00000007248.5
|
Hspa1l
|
heat shock protein 1-like |
| chr4_+_144619696 | 0.04 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr11_+_115310963 | 0.04 |
ENSMUST00000106533.8
ENSMUST00000123345.2 |
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr9_+_21279802 | 0.04 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr13_+_120151982 | 0.04 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr2_-_155953874 | 0.04 |
ENSMUST00000059647.12
ENSMUST00000109604.9 ENSMUST00000138068.2 ENSMUST00000128499.2 ENSMUST00000142960.9 ENSMUST00000136296.9 |
Rbm12
Cpne1
|
RNA binding motif protein 12 copine I |
| chr7_+_127345909 | 0.04 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr17_-_81035453 | 0.04 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr19_+_41471395 | 0.04 |
ENSMUST00000237208.2
ENSMUST00000238398.2 |
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr7_+_89779564 | 0.04 |
ENSMUST00000208742.2
ENSMUST00000049537.9 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_168259465 | 0.04 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr13_+_58956495 | 0.04 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr18_-_47501897 | 0.04 |
ENSMUST00000019791.14
ENSMUST00000115449.9 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_-_152673032 | 0.04 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr7_+_127084283 | 0.03 |
ENSMUST00000048896.8
|
Fbrs
|
fibrosin |
| chrX_+_47430221 | 0.03 |
ENSMUST00000136348.8
|
Bcorl1
|
BCL6 co-repressor-like 1 |
| chr5_+_110283689 | 0.03 |
ENSMUST00000200038.2
ENSMUST00000112519.9 ENSMUST00000198633.5 ENSMUST00000014812.13 ENSMUST00000199811.3 |
Chfr
|
checkpoint with forkhead and ring finger domains |
| chr7_-_46445305 | 0.03 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr9_+_21279179 | 0.03 |
ENSMUST00000213518.2
ENSMUST00000216892.2 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr7_+_16135112 | 0.03 |
ENSMUST00000098789.5
ENSMUST00000209289.2 |
Zc3h4
|
zinc finger CCCH-type containing 4 |
| chr10_-_68114543 | 0.03 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr14_-_54878804 | 0.03 |
ENSMUST00000067784.8
|
Cdh24
|
cadherin-like 24 |
| chr6_+_17463925 | 0.03 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr3_-_94490023 | 0.03 |
ENSMUST00000029783.16
|
Snx27
|
sorting nexin family member 27 |
| chr3_+_104696342 | 0.03 |
ENSMUST00000106787.8
ENSMUST00000176347.6 |
Rhoc
|
ras homolog family member C |
| chr9_+_56326089 | 0.03 |
ENSMUST00000213242.2
ENSMUST00000214771.2 ENSMUST00000217518.2 ENSMUST00000214869.2 |
Hmg20a
|
high mobility group 20A |
| chr2_-_75534985 | 0.03 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr14_-_103336391 | 0.03 |
ENSMUST00000132004.8
|
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr4_-_148215135 | 0.03 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr11_+_69909245 | 0.03 |
ENSMUST00000231415.2
ENSMUST00000108588.9 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr13_+_118851214 | 0.03 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr13_+_58956077 | 0.03 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr17_-_85097945 | 0.02 |
ENSMUST00000112308.9
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr9_+_95441652 | 0.02 |
ENSMUST00000079597.7
|
Paqr9
|
progestin and adipoQ receptor family member IX |
| chrX_-_133909934 | 0.02 |
ENSMUST00000113185.9
ENSMUST00000064659.6 |
Zmat1
|
zinc finger, matrin type 1 |
| chr11_+_101358990 | 0.02 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr1_-_84816379 | 0.02 |
ENSMUST00000187818.2
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_-_119308094 | 0.02 |
ENSMUST00000110808.2
ENSMUST00000049920.14 |
Ino80
|
INO80 complex subunit |
| chr15_-_50753437 | 0.02 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr2_-_73722932 | 0.02 |
ENSMUST00000154456.8
ENSMUST00000090802.11 ENSMUST00000055833.12 ENSMUST00000112007.8 ENSMUST00000112016.9 |
Atf2
|
activating transcription factor 2 |
| chr11_+_115310885 | 0.02 |
ENSMUST00000103035.10
|
Kctd2
|
potassium channel tetramerisation domain containing 2 |
| chr6_+_30048061 | 0.02 |
ENSMUST00000115209.8
ENSMUST00000115200.8 ENSMUST00000115204.8 ENSMUST00000148990.8 |
Nrf1
|
nuclear respiratory factor 1 |
| chr14_-_103336427 | 0.02 |
ENSMUST00000145693.8
|
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr11_+_100510043 | 0.02 |
ENSMUST00000107376.8
|
Nkiras2
|
NFKB inhibitor interacting Ras-like protein 2 |
| chr3_+_104696108 | 0.02 |
ENSMUST00000002303.12
|
Rhoc
|
ras homolog family member C |
| chr9_-_110571645 | 0.02 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr1_-_64160557 | 0.02 |
ENSMUST00000055001.10
ENSMUST00000114086.8 |
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_-_72876686 | 0.02 |
ENSMUST00000206378.2
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr11_+_3239868 | 0.02 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr13_-_103911092 | 0.02 |
ENSMUST00000074616.7
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr6_-_72876882 | 0.02 |
ENSMUST00000068697.11
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr4_+_137720755 | 0.02 |
ENSMUST00000084214.12
ENSMUST00000105831.9 ENSMUST00000203828.3 |
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr9_-_108183140 | 0.02 |
ENSMUST00000195615.2
|
Tcta
|
T cell leukemia translocation altered gene |
| chr9_+_56325893 | 0.01 |
ENSMUST00000034879.5
ENSMUST00000215269.2 |
Hmg20a
|
high mobility group 20A |
| chr15_+_102315579 | 0.01 |
ENSMUST00000169619.2
|
Sp1
|
trans-acting transcription factor 1 |
| chr13_+_83720457 | 0.01 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr3_-_142587419 | 0.01 |
ENSMUST00000174422.8
ENSMUST00000173830.8 |
Pkn2
|
protein kinase N2 |
| chr4_+_144619647 | 0.01 |
ENSMUST00000154208.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr7_-_19504446 | 0.01 |
ENSMUST00000003061.14
|
Bcam
|
basal cell adhesion molecule |
| chr15_+_89383799 | 0.01 |
ENSMUST00000109309.9
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr6_+_17463748 | 0.01 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr8_+_111782951 | 0.01 |
ENSMUST00000210390.2
|
Exosc6
|
exosome component 6 |
| chr13_+_38009951 | 0.01 |
ENSMUST00000138043.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr7_-_125799570 | 0.01 |
ENSMUST00000009344.16
|
Xpo6
|
exportin 6 |
| chr2_-_19002932 | 0.01 |
ENSMUST00000006912.12
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr7_-_109271171 | 0.01 |
ENSMUST00000208734.2
|
Denn2b
|
DENN domain containing 2B |
| chr11_+_96162283 | 0.01 |
ENSMUST00000000010.9
ENSMUST00000174042.3 |
Hoxb9
|
homeobox B9 |
| chrX_-_93166992 | 0.01 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr11_-_88609048 | 0.01 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr13_+_119565669 | 0.01 |
ENSMUST00000173627.8
ENSMUST00000126957.9 ENSMUST00000174691.8 |
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr13_+_38010203 | 0.01 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr13_+_119565424 | 0.01 |
ENSMUST00000026520.14
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr11_+_115921129 | 0.01 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr13_+_120151915 | 0.01 |
ENSMUST00000225543.2
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr10_-_120312374 | 0.01 |
ENSMUST00000072777.14
ENSMUST00000159699.2 |
Hmga2
|
high mobility group AT-hook 2 |
| chr14_-_103336990 | 0.01 |
ENSMUST00000022720.15
ENSMUST00000144141.8 |
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr15_+_89384317 | 0.01 |
ENSMUST00000135214.2
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr4_+_119112692 | 0.00 |
ENSMUST00000094823.4
|
Cldn19
|
claudin 19 |
| chr11_-_102187445 | 0.00 |
ENSMUST00000107132.3
ENSMUST00000073234.9 |
Atxn7l3
|
ataxin 7-like 3 |
| chr13_-_63721412 | 0.00 |
ENSMUST00000195106.2
|
Ptch1
|
patched 1 |
| chr5_-_29683468 | 0.00 |
ENSMUST00000165512.4
ENSMUST00000001608.8 |
Mnx1
|
motor neuron and pancreas homeobox 1 |
| chr19_-_46314945 | 0.00 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr7_-_142211203 | 0.00 |
ENSMUST00000097936.9
ENSMUST00000000033.12 |
Igf2
|
insulin-like growth factor 2 |
| chr13_+_48414582 | 0.00 |
ENSMUST00000021810.3
|
Id4
|
inhibitor of DNA binding 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Glis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:2000019 | positive regulation of dermatome development(GO:0061184) renal vesicle induction(GO:0072034) negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0002658 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0045212 | neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |