Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
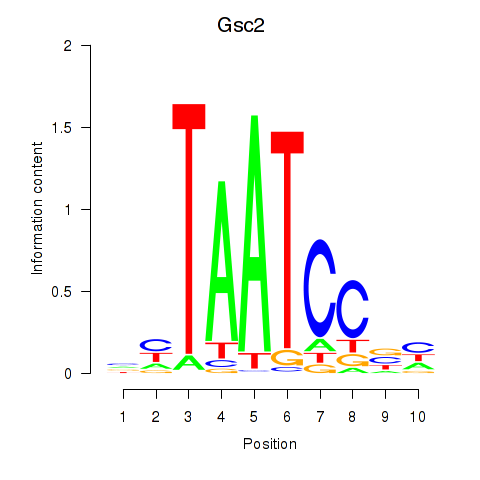
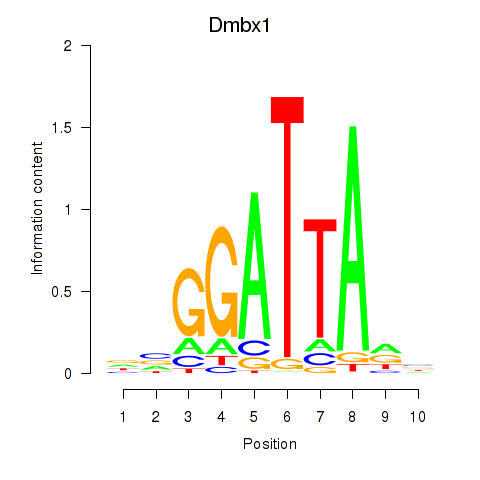
Results for Gsc2_Dmbx1
Z-value: 0.88
Transcription factors associated with Gsc2_Dmbx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsc2
|
ENSMUSG00000022738.8 | goosecoid homebox 2 |
|
Dmbx1
|
ENSMUSG00000028707.16 | diencephalon/mesencephalon homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmbx1 | mm39_v1_chr4_-_115797118_115797129 | 0.88 | 5.1e-02 | Click! |
| Gsc2 | mm39_v1_chr16_-_17732923_17732923 | 0.61 | 2.7e-01 | Click! |
Activity profile of Gsc2_Dmbx1 motif
Sorted Z-values of Gsc2_Dmbx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_44258112 | 1.82 |
ENSMUST00000054801.4
|
Mettl21e
|
methyltransferase like 21E |
| chr8_+_13455080 | 1.42 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chr13_+_23936250 | 1.38 |
ENSMUST00000091703.3
|
H3c2
|
H3 clustered histone 2 |
| chr13_-_21967540 | 0.87 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr6_+_136931519 | 0.71 |
ENSMUST00000137768.2
|
Pde6h
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr14_+_99536111 | 0.70 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5 |
| chr8_+_117822593 | 0.67 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr11_-_70350783 | 0.53 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr11_-_5049082 | 0.47 |
ENSMUST00000063232.7
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr9_+_50528813 | 0.42 |
ENSMUST00000141366.8
|
Pih1d2
|
PIH1 domain containing 2 |
| chr9_-_50528530 | 0.40 |
ENSMUST00000147671.2
ENSMUST00000145139.8 ENSMUST00000155435.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr3_-_103645311 | 0.37 |
ENSMUST00000029440.10
|
Olfml3
|
olfactomedin-like 3 |
| chr4_+_84802592 | 0.36 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein |
| chr6_+_3993774 | 0.35 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr10_-_22025169 | 0.32 |
ENSMUST00000179054.8
ENSMUST00000069372.7 |
E030030I06Rik
|
RIKEN cDNA E030030I06 gene |
| chr14_-_20027279 | 0.32 |
ENSMUST00000160013.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_-_84817976 | 0.31 |
ENSMUST00000190067.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr7_-_98305737 | 0.28 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr19_-_4189603 | 0.27 |
ENSMUST00000025761.8
|
Cabp4
|
calcium binding protein 4 |
| chr2_+_101455022 | 0.27 |
ENSMUST00000044031.4
|
Rag2
|
recombination activating gene 2 |
| chr10_+_128061699 | 0.25 |
ENSMUST00000026455.8
|
Mip
|
major intrinsic protein of lens fiber |
| chr14_+_28740162 | 0.25 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr4_+_136349924 | 0.24 |
ENSMUST00000178843.3
|
Lactbl1
|
lactamase, beta-like 1 |
| chrX_+_159551009 | 0.24 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr9_+_30941924 | 0.24 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr7_-_98305986 | 0.23 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr5_+_77413282 | 0.22 |
ENSMUST00000080359.12
|
Rest
|
RE1-silencing transcription factor |
| chr11_-_5049036 | 0.21 |
ENSMUST00000102930.10
ENSMUST00000093365.12 ENSMUST00000073308.11 |
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr11_-_51647290 | 0.20 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr17_+_36172235 | 0.18 |
ENSMUST00000172931.2
|
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr1_-_119576632 | 0.18 |
ENSMUST00000163147.8
ENSMUST00000052404.13 ENSMUST00000191046.7 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_+_178014983 | 0.18 |
ENSMUST00000161075.8
ENSMUST00000027783.14 |
Desi2
|
desumoylating isopeptidase 2 |
| chr2_+_121287444 | 0.17 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chr7_+_13132040 | 0.17 |
ENSMUST00000005791.14
|
Cabp5
|
calcium binding protein 5 |
| chr7_-_126574959 | 0.17 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr2_-_29983618 | 0.17 |
ENSMUST00000081838.7
ENSMUST00000102865.11 |
Zdhhc12
|
zinc finger, DHHC domain containing 12 |
| chr11_-_5049223 | 0.16 |
ENSMUST00000079949.13
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr11_-_69127848 | 0.15 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr6_-_29380467 | 0.14 |
ENSMUST00000080428.13
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr17_+_36172210 | 0.14 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr1_-_13730732 | 0.13 |
ENSMUST00000027071.7
|
Lactb2
|
lactamase, beta 2 |
| chr4_+_126503611 | 0.12 |
ENSMUST00000097886.4
ENSMUST00000164362.2 |
5730409E04Rik
|
RIKEN cDNA 5730409E04Rik gene |
| chr7_-_127324788 | 0.12 |
ENSMUST00000076091.4
|
Ctf2
|
cardiotrophin 2 |
| chr3_-_88368489 | 0.12 |
ENSMUST00000166237.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_-_52173391 | 0.12 |
ENSMUST00000086844.10
|
Tcf7
|
transcription factor 7, T cell specific |
| chr7_-_4755971 | 0.12 |
ENSMUST00000183971.8
ENSMUST00000182173.2 ENSMUST00000182738.8 ENSMUST00000182111.8 ENSMUST00000184143.8 ENSMUST00000182048.2 ENSMUST00000063324.14 |
Cox6b2
|
cytochrome c oxidase subunit 6B2 |
| chr8_-_96033213 | 0.12 |
ENSMUST00000119870.9
ENSMUST00000093268.5 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr7_+_101619327 | 0.12 |
ENSMUST00000210475.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr2_-_22930149 | 0.10 |
ENSMUST00000091394.13
ENSMUST00000093171.13 |
Abi1
|
abl interactor 1 |
| chr7_-_105401398 | 0.10 |
ENSMUST00000033184.6
|
Tpp1
|
tripeptidyl peptidase I |
| chr18_-_46730547 | 0.10 |
ENSMUST00000151189.2
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr11_-_70510010 | 0.10 |
ENSMUST00000102556.10
ENSMUST00000014753.9 |
Chrne
|
cholinergic receptor, nicotinic, epsilon polypeptide |
| chr7_+_57069417 | 0.09 |
ENSMUST00000085240.11
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 |
| chr1_-_132294807 | 0.08 |
ENSMUST00000136828.3
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr5_-_31359276 | 0.08 |
ENSMUST00000031562.11
|
Zfp513
|
zinc finger protein 513 |
| chr19_+_26582450 | 0.08 |
ENSMUST00000176769.9
ENSMUST00000208163.2 ENSMUST00000025862.15 ENSMUST00000176030.8 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_101619019 | 0.07 |
ENSMUST00000084852.13
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr17_-_35383867 | 0.07 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr6_+_115111872 | 0.06 |
ENSMUST00000009538.12
ENSMUST00000203450.2 |
Syn2
|
synapsin II |
| chr6_-_124817155 | 0.06 |
ENSMUST00000024206.6
|
Gnb3
|
guanine nucleotide binding protein (G protein), beta 3 |
| chr11_+_5049608 | 0.06 |
ENSMUST00000139742.8
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr3_+_28835425 | 0.05 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr19_+_4189786 | 0.05 |
ENSMUST00000096338.5
|
Gpr152
|
G protein-coupled receptor 152 |
| chr7_+_103684652 | 0.04 |
ENSMUST00000213214.2
|
Olfr641
|
olfactory receptor 641 |
| chr2_-_73316809 | 0.04 |
ENSMUST00000141264.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr15_-_65848649 | 0.03 |
ENSMUST00000239034.2
ENSMUST00000100584.3 |
Hhla1
|
HERV-H LTR-associating 1 |
| chr11_+_67586140 | 0.03 |
ENSMUST00000021290.2
|
Rcvrn
|
recoverin |
| chr19_+_8595369 | 0.03 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr7_-_132616977 | 0.03 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr8_-_106863423 | 0.03 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr17_-_85397627 | 0.03 |
ENSMUST00000072406.5
ENSMUST00000171795.9 ENSMUST00000234676.2 |
Prepl
|
prolyl endopeptidase-like |
| chr3_-_84167119 | 0.02 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr11_+_70350963 | 0.02 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr17_-_35265514 | 0.02 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr7_+_43361930 | 0.02 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr7_-_80053063 | 0.02 |
ENSMUST00000147150.2
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_+_32305259 | 0.02 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr11_+_117090465 | 0.02 |
ENSMUST00000093907.11
|
Septin9
|
septin 9 |
| chr14_-_55762416 | 0.02 |
ENSMUST00000178694.3
|
Nrl
|
neural retina leucine zipper gene |
| chr14_+_36776775 | 0.01 |
ENSMUST00000120052.2
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr6_-_29380423 | 0.01 |
ENSMUST00000147483.3
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) |
| chr14_-_55762432 | 0.01 |
ENSMUST00000062232.15
|
Nrl
|
neural retina leucine zipper gene |
| chr16_-_17732923 | 0.01 |
ENSMUST00000012279.6
ENSMUST00000232493.2 |
Gsc2
|
goosecoid homebox 2 |
| chr4_-_137493785 | 0.01 |
ENSMUST00000139951.8
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr11_-_99176086 | 0.01 |
ENSMUST00000017255.4
|
Krt24
|
keratin 24 |
| chr1_+_82817794 | 0.01 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr7_-_132725575 | 0.01 |
ENSMUST00000171968.8
|
Ctbp2
|
C-terminal binding protein 2 |
| chr11_-_90578397 | 0.01 |
ENSMUST00000107869.9
ENSMUST00000154599.2 ENSMUST00000107868.8 ENSMUST00000020849.9 |
Tom1l1
|
target of myb1-like 1 (chicken) |
| chr11_+_3981769 | 0.01 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr11_+_70350725 | 0.01 |
ENSMUST00000147289.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr19_-_47680528 | 0.01 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr1_-_155108455 | 0.01 |
ENSMUST00000035914.5
|
BC034090
|
cDNA sequence BC034090 |
| chr19_-_13080648 | 0.01 |
ENSMUST00000076729.4
|
Olfr1458
|
olfactory receptor 1458 |
| chr13_-_64645606 | 0.01 |
ENSMUST00000180282.2
|
Fam240b
|
family with sequence similarity 240 member B |
| chr19_-_46314945 | 0.01 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr11_-_43792013 | 0.01 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr5_-_137643954 | 0.01 |
ENSMUST00000057314.4
|
Irs3
|
insulin receptor substrate 3 |
| chr16_+_17051423 | 0.01 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr4_-_140781015 | 0.01 |
ENSMUST00000102491.10
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr4_-_139974062 | 0.01 |
ENSMUST00000039331.9
|
Igsf21
|
immunoglobulin superfamily, member 21 |
| chr1_-_154692678 | 0.01 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr5_-_66161621 | 0.00 |
ENSMUST00000201351.4
|
9130230L23Rik
|
RIKEN cDNA 9130230L23 gene |
| chr13_-_113755082 | 0.00 |
ENSMUST00000109241.5
|
Snx18
|
sorting nexin 18 |
| chr2_+_22958956 | 0.00 |
ENSMUST00000226571.2
ENSMUST00000114529.10 ENSMUST00000114526.9 ENSMUST00000228050.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr17_+_47221377 | 0.00 |
ENSMUST00000024773.6
|
Prph2
|
peripherin 2 |
| chr1_+_178015287 | 0.00 |
ENSMUST00000159284.2
|
Desi2
|
desumoylating isopeptidase 2 |
| chr2_+_164404499 | 0.00 |
ENSMUST00000017867.10
ENSMUST00000109344.9 ENSMUST00000109345.9 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chrX_+_133195974 | 0.00 |
ENSMUST00000037687.8
|
Tmem35a
|
transmembrane protein 35A |
| chr4_-_139560831 | 0.00 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr16_-_29197485 | 0.00 |
ENSMUST00000143373.8
ENSMUST00000075806.11 |
Atp13a5
|
ATPase type 13A5 |
| chrX_+_132809166 | 0.00 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_+_43418404 | 0.00 |
ENSMUST00000014063.6
|
Klk12
|
kallikrein related-peptidase 12 |
| chr17_-_72059865 | 0.00 |
ENSMUST00000057405.9
|
Pcare
|
photoreceptor cilium actin regulator |
| chr7_+_63803583 | 0.00 |
ENSMUST00000085222.12
ENSMUST00000206277.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_+_63803663 | 0.00 |
ENSMUST00000206314.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr19_+_12861219 | 0.00 |
ENSMUST00000049624.3
|
Olfr1445
|
olfactory receptor 1445 |
| chr6_-_38086484 | 0.00 |
ENSMUST00000114908.5
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr7_+_12246415 | 0.00 |
ENSMUST00000032541.5
|
2900092C05Rik
|
RIKEN cDNA 2900092C05 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsc2_Dmbx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.2 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) B cell homeostatic proliferation(GO:0002358) |
| 0.0 | 0.2 | GO:1902365 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.1 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |