Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
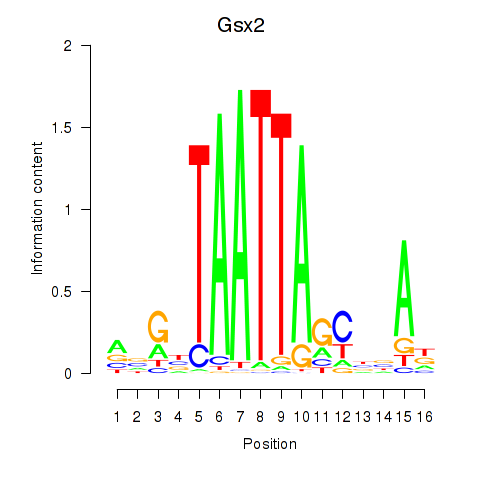
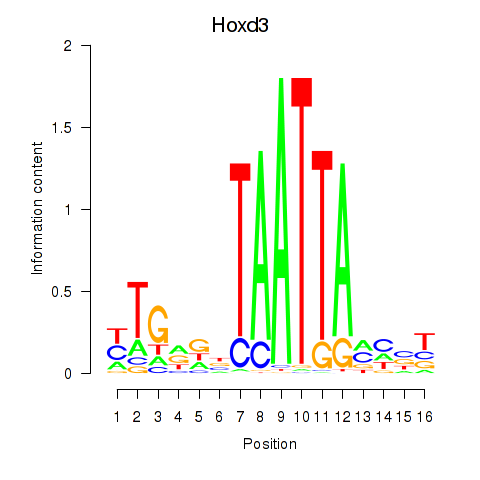
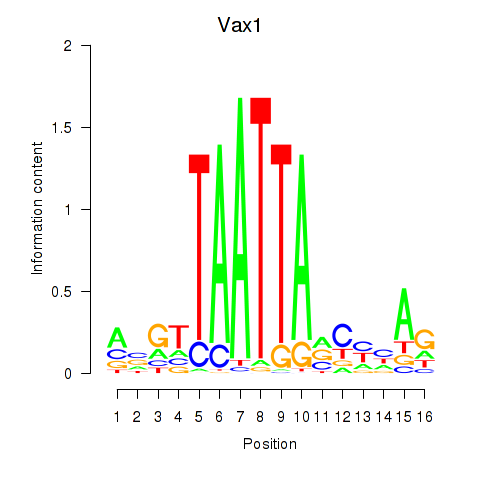
Results for Gsx2_Hoxd3_Vax1
Z-value: 0.16
Transcription factors associated with Gsx2_Hoxd3_Vax1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gsx2
|
ENSMUSG00000035946.8 | GS homeobox 2 |
|
Hoxd3
|
ENSMUSG00000079277.10 | homeobox D3 |
|
Vax1
|
ENSMUSG00000006270.8 | ventral anterior homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxd3 | mm39_v1_chr2_+_74542255_74542283 | 0.62 | 2.7e-01 | Click! |
| Gsx2 | mm39_v1_chr5_+_75236250_75236263 | 0.39 | 5.1e-01 | Click! |
Activity profile of Gsx2_Hoxd3_Vax1 motif
Sorted Z-values of Gsx2_Hoxd3_Vax1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_118892497 | 0.42 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chrM_+_9870 | 0.15 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr4_+_8690398 | 0.09 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr8_+_66838927 | 0.09 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chrM_+_14138 | 0.08 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.08 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr7_-_3901119 | 0.08 |
ENSMUST00000070639.8
|
Gm14548
|
predicted gene 14548 |
| chr2_+_91480513 | 0.08 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr14_-_50586329 | 0.07 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr2_-_111820618 | 0.07 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr15_-_77840856 | 0.07 |
ENSMUST00000117725.2
ENSMUST00000016696.13 |
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr2_-_111843053 | 0.07 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr17_+_17622934 | 0.07 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr13_+_22656093 | 0.07 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr5_-_84565218 | 0.06 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr13_+_22508759 | 0.06 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr4_-_43710231 | 0.06 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr9_+_40092216 | 0.06 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr5_-_137015683 | 0.06 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr17_+_85335775 | 0.06 |
ENSMUST00000024944.9
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr11_-_109502243 | 0.05 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr7_-_63586049 | 0.05 |
ENSMUST00000185175.2
|
Klf13
|
Kruppel-like factor 13 |
| chr2_-_45007407 | 0.05 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_159551009 | 0.05 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr2_+_91480460 | 0.05 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr16_-_16418397 | 0.04 |
ENSMUST00000159542.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr19_+_8975249 | 0.04 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr3_-_129834788 | 0.04 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae) |
| chr9_+_39932760 | 0.04 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr1_+_88139678 | 0.04 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr7_-_123099672 | 0.04 |
ENSMUST00000042470.14
ENSMUST00000128217.2 |
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr16_+_51851948 | 0.04 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_-_12829100 | 0.04 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr7_-_11414074 | 0.04 |
ENSMUST00000227010.2
ENSMUST00000209638.3 |
Vmn1r72
|
vomeronasal 1 receptor 72 |
| chr2_-_111880531 | 0.04 |
ENSMUST00000213582.2
ENSMUST00000213961.3 ENSMUST00000215531.2 |
Olfr1312
|
olfactory receptor 1312 |
| chr3_-_15902583 | 0.04 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr12_-_99529767 | 0.04 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr7_+_108265625 | 0.04 |
ENSMUST00000213979.3
ENSMUST00000216331.2 ENSMUST00000217170.2 |
Olfr510
|
olfactory receptor 510 |
| chr3_-_15640045 | 0.04 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr5_+_104350475 | 0.03 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr1_-_92412835 | 0.03 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr6_-_116084810 | 0.03 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr6_+_54794433 | 0.03 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr7_-_10292412 | 0.03 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr16_-_4698148 | 0.03 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr4_-_15149755 | 0.03 |
ENSMUST00000108273.2
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr15_+_25774070 | 0.03 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr16_+_51851917 | 0.03 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr12_+_112727089 | 0.03 |
ENSMUST00000063888.5
|
Pld4
|
phospholipase D family, member 4 |
| chr9_+_20193647 | 0.03 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr2_-_45002902 | 0.03 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr13_-_53627110 | 0.03 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr3_+_89153258 | 0.03 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr9_+_110306052 | 0.03 |
ENSMUST00000197248.5
ENSMUST00000061155.12 ENSMUST00000198043.5 ENSMUST00000084952.8 |
Kif9
|
kinesin family member 9 |
| chr5_+_137015873 | 0.03 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr11_-_73382303 | 0.02 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr4_-_148236516 | 0.02 |
ENSMUST00000056965.12
ENSMUST00000168503.8 ENSMUST00000152098.8 |
Fbxo6
|
F-box protein 6 |
| chr16_-_89615225 | 0.02 |
ENSMUST00000164263.9
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr2_-_27365633 | 0.02 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chrM_+_10167 | 0.02 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr5_+_87814058 | 0.02 |
ENSMUST00000199506.5
ENSMUST00000197631.5 ENSMUST00000094641.9 |
Csn1s1
|
casein alpha s1 |
| chr5_+_138185747 | 0.02 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr1_+_130754413 | 0.02 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr16_+_51851588 | 0.02 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr6_-_36787096 | 0.02 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr3_-_64473251 | 0.02 |
ENSMUST00000176481.9
|
Vmn2r6
|
vomeronasal 2, receptor 6 |
| chr6_-_123395075 | 0.02 |
ENSMUST00000172199.3
|
Vmn2r20
|
vomeronasal 2, receptor 20 |
| chr12_+_52746158 | 0.02 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_-_87742525 | 0.02 |
ENSMUST00000164517.3
|
Eif1ad19
|
eukaryotic translation initiation factor 1A domain containing 19 |
| chr2_-_85632888 | 0.02 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr17_-_47813201 | 0.02 |
ENSMUST00000233174.2
ENSMUST00000233121.2 ENSMUST00000067103.4 |
Taf8
|
TATA-box binding protein associated factor 8 |
| chr4_+_100336003 | 0.02 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_-_102507962 | 0.02 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr14_-_31139617 | 0.02 |
ENSMUST00000100730.10
|
Sh3bp5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr7_+_43885573 | 0.02 |
ENSMUST00000223070.2
ENSMUST00000205530.2 |
Gm36864
|
predicted gene, 36864 |
| chr3_-_88204286 | 0.02 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr5_-_137101108 | 0.02 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr17_-_21006419 | 0.02 |
ENSMUST00000233605.2
ENSMUST00000232812.2 ENSMUST00000233186.2 ENSMUST00000233164.2 |
Vmn1r228
|
vomeronasal 1 receptor 228 |
| chr3_-_131096792 | 0.02 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr13_-_23041731 | 0.02 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr10_+_81229443 | 0.02 |
ENSMUST00000118206.2
|
Smim24
|
small integral membrane protein 24 |
| chr2_+_69727599 | 0.02 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_+_87696836 | 0.02 |
ENSMUST00000213308.3
|
Olfr1152
|
olfactory receptor 1152 |
| chr1_+_75412574 | 0.02 |
ENSMUST00000037796.14
ENSMUST00000113584.8 ENSMUST00000145166.8 ENSMUST00000143730.8 ENSMUST00000133418.8 ENSMUST00000144874.8 ENSMUST00000140287.8 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chrX_-_156198282 | 0.02 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr9_+_121780054 | 0.02 |
ENSMUST00000043011.9
ENSMUST00000214536.3 ENSMUST00000215990.3 |
Gask1a
|
golgi associated kinase 1A |
| chr5_-_100563974 | 0.02 |
ENSMUST00000182886.8
ENSMUST00000094578.11 |
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr10_-_81243475 | 0.02 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr10_-_75946790 | 0.02 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr9_+_32027335 | 0.01 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr9_-_110305705 | 0.01 |
ENSMUST00000198164.5
ENSMUST00000068025.13 |
Klhl18
|
kelch-like 18 |
| chr10_-_129965752 | 0.01 |
ENSMUST00000215217.2
ENSMUST00000214192.2 |
Olfr824
|
olfactory receptor 824 |
| chr5_-_3697806 | 0.01 |
ENSMUST00000119783.2
ENSMUST00000007559.15 |
Gatad1
|
GATA zinc finger domain containing 1 |
| chr18_+_12874390 | 0.01 |
ENSMUST00000121018.8
ENSMUST00000119108.8 ENSMUST00000186263.2 ENSMUST00000191078.7 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr2_-_88157559 | 0.01 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr8_+_84262409 | 0.01 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr6_-_125357756 | 0.01 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr16_-_19341016 | 0.01 |
ENSMUST00000214315.2
|
Olfr167
|
olfactory receptor 167 |
| chr17_+_38104635 | 0.01 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr10_+_34265969 | 0.01 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr7_+_108533613 | 0.01 |
ENSMUST00000033342.7
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr13_-_22689551 | 0.01 |
ENSMUST00000228020.2
|
Vmn1r202
|
vomeronasal 1 receptor 202 |
| chr1_+_78286946 | 0.01 |
ENSMUST00000036172.10
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr17_+_38104420 | 0.01 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr10_+_62860291 | 0.01 |
ENSMUST00000020262.5
|
Pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr1_-_69726384 | 0.01 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr2_+_88470886 | 0.01 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr17_+_21031817 | 0.01 |
ENSMUST00000232810.2
ENSMUST00000233712.2 ENSMUST00000232852.2 |
Vmn1r229
|
vomeronasal 1 receptor 229 |
| chr10_-_128875765 | 0.01 |
ENSMUST00000204763.3
|
Olfr764-ps1
|
olfactory receptor 764, pseudogene 1 |
| chr8_-_41494890 | 0.01 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr16_+_45044678 | 0.01 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr3_+_53396120 | 0.01 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr7_-_102540089 | 0.01 |
ENSMUST00000217024.2
|
Olfr569
|
olfactory receptor 569 |
| chr9_+_27210500 | 0.01 |
ENSMUST00000214357.2
ENSMUST00000115247.8 ENSMUST00000133213.3 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr6_-_30693675 | 0.01 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr5_-_62923463 | 0.01 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr2_-_85966272 | 0.01 |
ENSMUST00000216566.3
ENSMUST00000214364.2 |
Olfr1039
|
olfactory receptor 1039 |
| chr6_-_69704122 | 0.01 |
ENSMUST00000103364.3
|
Igkv5-48
|
immunoglobulin kappa variable 5-48 |
| chr7_+_126808016 | 0.01 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_-_73217633 | 0.01 |
ENSMUST00000134079.2
|
Aspa
|
aspartoacylase |
| chr11_+_58549642 | 0.01 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr9_-_107749594 | 0.01 |
ENSMUST00000183035.2
|
Rbm6
|
RNA binding motif protein 6 |
| chr7_+_84502761 | 0.01 |
ENSMUST00000217039.3
ENSMUST00000211582.2 |
Olfr291
|
olfactory receptor 291 |
| chr11_-_73217298 | 0.01 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr3_-_58729732 | 0.01 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr2_+_177760768 | 0.01 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr19_-_5610628 | 0.01 |
ENSMUST00000025861.3
|
Ovol1
|
ovo like zinc finger 1 |
| chr7_-_102638531 | 0.01 |
ENSMUST00000215606.2
|
Olfr578
|
olfactory receptor 578 |
| chr7_+_10365459 | 0.01 |
ENSMUST00000226255.2
ENSMUST00000228090.2 |
Vmn1r70
|
vomeronasal 1 receptor 70 |
| chr4_+_145397238 | 0.01 |
ENSMUST00000105738.9
|
Zfp980
|
zinc finger protein 980 |
| chr11_-_65160767 | 0.01 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr11_-_65160810 | 0.01 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr4_+_108576846 | 0.01 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr2_+_88505972 | 0.01 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr2_-_168609110 | 0.01 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chr4_+_147637714 | 0.01 |
ENSMUST00000139784.8
ENSMUST00000143885.8 ENSMUST00000081742.7 |
Zfp985
|
zinc finger protein 985 |
| chr4_+_145311759 | 0.01 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr8_-_87307294 | 0.01 |
ENSMUST00000131423.8
ENSMUST00000152438.2 |
Abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr5_-_143279378 | 0.01 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr11_-_99265721 | 0.01 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr11_-_79418500 | 0.01 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr2_-_164013033 | 0.00 |
ENSMUST00000045196.4
|
Kcns1
|
K+ voltage-gated channel, subfamily S, 1 |
| chr15_-_8740218 | 0.00 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr9_+_38725910 | 0.00 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chr10_+_119828867 | 0.00 |
ENSMUST00000154238.8
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr7_+_45271229 | 0.00 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr9_-_103099262 | 0.00 |
ENSMUST00000170904.2
|
Trf
|
transferrin |
| chr4_+_133280680 | 0.00 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr2_+_164455438 | 0.00 |
ENSMUST00000094346.3
|
Wfdc6b
|
WAP four-disulfide core domain 6B |
| chr6_+_37847721 | 0.00 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr16_+_88828587 | 0.00 |
ENSMUST00000076906.2
|
Krtap6-1
|
keratin associated protein 6-1 |
| chr10_+_119828821 | 0.00 |
ENSMUST00000105261.9
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr16_-_45544960 | 0.00 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr12_-_79343040 | 0.00 |
ENSMUST00000218377.2
ENSMUST00000021547.8 |
Zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr19_-_40371016 | 0.00 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr9_-_39465349 | 0.00 |
ENSMUST00000215505.2
ENSMUST00000217227.2 |
Olfr958
|
olfactory receptor 958 |
| chr2_-_91480096 | 0.00 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr18_+_12874368 | 0.00 |
ENSMUST00000235000.2
ENSMUST00000115857.9 |
Cabyr
|
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
| chr9_+_110306020 | 0.00 |
ENSMUST00000198858.5
|
Kif9
|
kinesin family member 9 |
| chr14_-_52704952 | 0.00 |
ENSMUST00000206520.3
|
Olfr1508
|
olfactory receptor 1508 |
| chrX_-_142716085 | 0.00 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr2_-_32977182 | 0.00 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr2_-_168608949 | 0.00 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr2_+_76480606 | 0.00 |
ENSMUST00000099986.3
|
Pjvk
|
pejvakin |
| chr18_+_46874920 | 0.00 |
ENSMUST00000025357.9
ENSMUST00000225520.2 |
Ap3s1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_+_55689921 | 0.00 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr1_+_54289833 | 0.00 |
ENSMUST00000027128.11
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr15_+_98468885 | 0.00 |
ENSMUST00000023728.8
|
Tex49
|
testis expressed 49 |
| chr15_-_82678490 | 0.00 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr1_-_37955569 | 0.00 |
ENSMUST00000078307.7
|
Lyg2
|
lysozyme G-like 2 |
| chr19_-_32173824 | 0.00 |
ENSMUST00000151822.2
|
Sgms1
|
sphingomyelin synthase 1 |
| chrX_+_105964224 | 0.00 |
ENSMUST00000060576.8
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr8_-_112737952 | 0.00 |
ENSMUST00000034426.14
|
Kars
|
lysyl-tRNA synthetase |
| chr7_+_4340708 | 0.00 |
ENSMUST00000006792.6
ENSMUST00000126417.3 |
Ncr1
|
natural cytotoxicity triggering receptor 1 |
| chr2_-_28806639 | 0.00 |
ENSMUST00000113847.3
|
Barhl1
|
BarH like homeobox 1 |
| chr17_+_88748139 | 0.00 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr4_+_146586445 | 0.00 |
ENSMUST00000105735.9
|
Zfp981
|
zinc finger protein 981 |
| chr11_+_73827503 | 0.00 |
ENSMUST00000214210.2
|
Olfr23
|
olfactory receptor 23 |
| chr11_-_73348284 | 0.00 |
ENSMUST00000121209.3
ENSMUST00000127789.3 |
Olfr380
|
olfactory receptor 380 |
| chr14_+_25980463 | 0.00 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr11_+_114742331 | 0.00 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chrX_-_36825123 | 0.00 |
ENSMUST00000115169.4
|
Rhox2g
|
reproductive homeobox 2G |
| chr11_+_100978103 | 0.00 |
ENSMUST00000107302.8
ENSMUST00000107303.10 ENSMUST00000017945.15 ENSMUST00000149597.2 |
Mlx
|
MAX-like protein X |
| chr9_+_38755745 | 0.00 |
ENSMUST00000217350.2
|
Olfr924
|
olfactory receptor 924 |
| chr19_+_57349112 | 0.00 |
ENSMUST00000036407.6
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr14_+_64229914 | 0.00 |
ENSMUST00000058229.6
|
Rp1l1
|
retinitis pigmentosa 1 homolog like 1 |
| chr2_-_164455520 | 0.00 |
ENSMUST00000094351.11
ENSMUST00000109338.2 |
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr2_-_89678487 | 0.00 |
ENSMUST00000214428.3
|
Olfr48
|
olfactory receptor 48 |
| chr19_-_7688628 | 0.00 |
ENSMUST00000025666.8
|
Slc22a19
|
solute carrier family 22 (organic anion transporter), member 19 |
| chr3_-_144680801 | 0.00 |
ENSMUST00000029923.10
ENSMUST00000238960.2 |
Clca4a
|
chloride channel accessory 4A |
| chr4_-_112632057 | 0.00 |
ENSMUST00000068851.6
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr7_-_8164653 | 0.00 |
ENSMUST00000168807.3
|
Vmn2r41
|
vomeronasal 2, receptor 41 |
| chr5_-_99091681 | 0.00 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr6_-_101354858 | 0.00 |
ENSMUST00000075994.11
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr11_+_114741948 | 0.00 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_+_106523532 | 0.00 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr3_-_144638284 | 0.00 |
ENSMUST00000098549.4
|
Clca4b
|
chloride channel accessory 4B |
| chr15_-_37459570 | 0.00 |
ENSMUST00000119730.8
ENSMUST00000120746.8 |
Ncald
|
neurocalcin delta |
| chr9_+_21437440 | 0.00 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr13_-_103042294 | 0.00 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Gsx2_Hoxd3_Vax1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:1903336 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.0 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |