Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
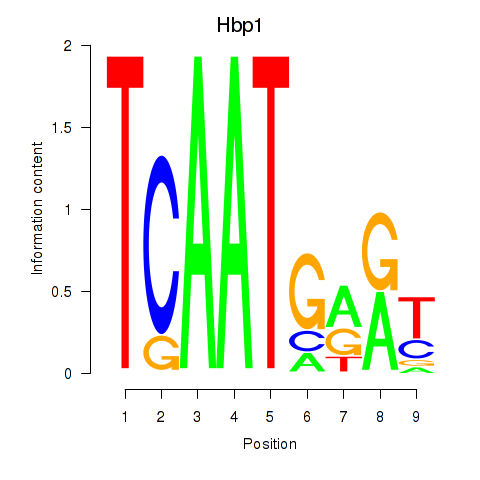
Results for Hbp1
Z-value: 0.98
Transcription factors associated with Hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hbp1
|
ENSMUSG00000002996.18 | high mobility group box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hbp1 | mm39_v1_chr12_-_32000534_32000558 | -0.84 | 7.8e-02 | Click! |
Activity profile of Hbp1 motif
Sorted Z-values of Hbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_84810802 | 0.86 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr1_-_140111138 | 0.65 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 0.60 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr17_+_31329939 | 0.33 |
ENSMUST00000236241.2
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chrX_+_93679671 | 0.33 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr14_-_79539063 | 0.32 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr8_+_120719177 | 0.32 |
ENSMUST00000132583.8
ENSMUST00000034282.16 |
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr1_+_88022776 | 0.32 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr1_+_87998487 | 0.31 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr7_+_16186704 | 0.30 |
ENSMUST00000019302.10
|
Tmem160
|
transmembrane protein 160 |
| chr13_+_21906214 | 0.29 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14 |
| chr11_-_87766350 | 0.29 |
ENSMUST00000049768.4
|
Epx
|
eosinophil peroxidase |
| chr9_+_123195986 | 0.27 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr2_-_151586063 | 0.26 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr5_-_23881353 | 0.24 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr1_+_87983099 | 0.24 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chrM_+_3906 | 0.22 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr13_+_21938258 | 0.22 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr7_-_19410749 | 0.22 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr13_-_23735822 | 0.22 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6 |
| chr17_+_29487881 | 0.22 |
ENSMUST00000234845.2
ENSMUST00000235038.2 ENSMUST00000235050.2 ENSMUST00000120346.9 ENSMUST00000234377.2 ENSMUST00000235074.2 ENSMUST00000235040.2 ENSMUST00000234256.2 ENSMUST00000234459.2 |
BC004004
|
cDNA sequence BC004004 |
| chr6_+_135339929 | 0.21 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr5_-_108022900 | 0.21 |
ENSMUST00000138111.8
ENSMUST00000112642.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr14_+_26722319 | 0.21 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr11_-_70242850 | 0.20 |
ENSMUST00000019068.7
|
Alox15
|
arachidonate 15-lipoxygenase |
| chrM_+_2743 | 0.20 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr9_-_58648826 | 0.19 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr9_-_49479791 | 0.19 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1 |
| chrM_+_7006 | 0.19 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr17_+_64907697 | 0.18 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr15_-_37004302 | 0.18 |
ENSMUST00000228275.2
|
Zfp706
|
zinc finger protein 706 |
| chr13_+_21901791 | 0.18 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr8_+_56747613 | 0.18 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr19_-_24938909 | 0.18 |
ENSMUST00000025815.10
|
Cbwd1
|
COBW domain containing 1 |
| chr13_-_23758370 | 0.17 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr3_-_130503041 | 0.17 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr3_-_144412394 | 0.16 |
ENSMUST00000200532.2
|
Sh3glb1
|
SH3-domain GRB2-like B1 (endophilin) |
| chr6_-_51446752 | 0.16 |
ENSMUST00000204188.3
ENSMUST00000203220.3 ENSMUST00000114459.8 ENSMUST00000090002.10 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr11_-_78642480 | 0.16 |
ENSMUST00000059468.6
|
Ccnq
|
cyclin Q |
| chr3_+_89173859 | 0.16 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr13_-_63036096 | 0.15 |
ENSMUST00000092888.11
|
Fbp1
|
fructose bisphosphatase 1 |
| chr19_-_4675352 | 0.15 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr1_-_69723316 | 0.15 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr17_+_29487762 | 0.15 |
ENSMUST00000064709.13
ENSMUST00000234711.2 |
BC004004
|
cDNA sequence BC004004 |
| chr7_-_43182595 | 0.15 |
ENSMUST00000205503.2
|
Cd33
|
CD33 antigen |
| chr11_+_51858476 | 0.15 |
ENSMUST00000102763.5
|
Cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr7_-_19411866 | 0.15 |
ENSMUST00000142352.9
|
Apoc2
|
apolipoprotein C-II |
| chr19_-_4675300 | 0.14 |
ENSMUST00000225264.2
ENSMUST00000237022.2 ENSMUST00000224675.3 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr11_-_106606076 | 0.14 |
ENSMUST00000080853.11
ENSMUST00000183610.8 ENSMUST00000103069.10 ENSMUST00000106796.9 |
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr15_-_102140331 | 0.14 |
ENSMUST00000230652.2
ENSMUST00000127014.3 ENSMUST00000001327.11 |
Itgb7
|
integrin beta 7 |
| chr7_-_43309563 | 0.14 |
ENSMUST00000032667.10
|
Siglece
|
sialic acid binding Ig-like lectin E |
| chr1_-_183766195 | 0.14 |
ENSMUST00000050306.8
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene |
| chr3_-_88317601 | 0.14 |
ENSMUST00000193338.6
ENSMUST00000056370.13 |
Pmf1
|
polyamine-modulated factor 1 |
| chr19_-_33495180 | 0.13 |
ENSMUST00000143522.9
|
Lipo4
|
lipase, member O4 |
| chrM_-_14061 | 0.13 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr8_-_34578880 | 0.13 |
ENSMUST00000080152.5
|
Gm10131
|
predicted pseudogene 10131 |
| chr11_+_46295547 | 0.13 |
ENSMUST00000063166.6
|
Fam71b
|
family with sequence similarity 71, member B |
| chr10_-_100425067 | 0.13 |
ENSMUST00000218821.2
ENSMUST00000054471.10 |
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr19_+_24853039 | 0.13 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chrM_+_14138 | 0.13 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr5_-_100710702 | 0.13 |
ENSMUST00000097437.9
|
Plac8
|
placenta-specific 8 |
| chr11_+_58197881 | 0.13 |
ENSMUST00000153510.9
|
Zfp692
|
zinc finger protein 692 |
| chr8_-_32440071 | 0.13 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chr7_+_28136861 | 0.13 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr7_+_127728712 | 0.13 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr4_-_148021217 | 0.12 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr5_-_124003553 | 0.12 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chrX_+_108240356 | 0.12 |
ENSMUST00000139259.2
ENSMUST00000060013.4 |
Gm6377
|
predicted gene 6377 |
| chr17_+_25946644 | 0.12 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr9_+_72946994 | 0.12 |
ENSMUST00000184126.3
|
Pigbos1
|
Pigb opposite strand 1 |
| chr3_-_105861497 | 0.12 |
ENSMUST00000199311.2
|
Atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr12_-_32111214 | 0.12 |
ENSMUST00000003079.12
ENSMUST00000036497.16 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr6_-_51446850 | 0.12 |
ENSMUST00000069949.13
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr10_+_80084955 | 0.12 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr8_-_46577183 | 0.12 |
ENSMUST00000170416.8
|
Snx25
|
sorting nexin 25 |
| chr9_+_55949141 | 0.12 |
ENSMUST00000114276.3
|
Rcn2
|
reticulocalbin 2 |
| chr15_+_78819378 | 0.12 |
ENSMUST00000145157.2
ENSMUST00000123013.2 |
Nol12
|
nucleolar protein 12 |
| chr18_-_25886750 | 0.11 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr17_+_31739089 | 0.11 |
ENSMUST00000064798.16
ENSMUST00000046288.16 ENSMUST00000191598.3 |
Ndufv3
|
NADH:ubiquinone oxidoreductase core subunit V3 |
| chr19_-_44017637 | 0.11 |
ENSMUST00000026211.10
ENSMUST00000211830.2 |
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr7_-_141934524 | 0.11 |
ENSMUST00000209263.2
|
Gm49369
|
predicted gene, 49369 |
| chr11_-_102360664 | 0.11 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr9_-_111086528 | 0.11 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chrM_+_11735 | 0.11 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr8_-_85751897 | 0.11 |
ENSMUST00000064314.10
|
Get3
|
guided entry of tail-anchored proteins factor 3, ATPase |
| chr4_+_148985584 | 0.11 |
ENSMUST00000147270.2
|
Casz1
|
castor zinc finger 1 |
| chr11_-_115310743 | 0.11 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr19_-_53577499 | 0.11 |
ENSMUST00000095978.5
|
Nutf2-ps1
|
nuclear transport factor 2, pseudogene 1 |
| chr5_-_110987604 | 0.11 |
ENSMUST00000056937.12
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr6_-_70149254 | 0.11 |
ENSMUST00000197272.2
|
Igkv8-27
|
immunoglobulin kappa chain variable 8-27 |
| chr9_-_106769069 | 0.10 |
ENSMUST00000160503.4
ENSMUST00000159620.9 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr5_-_151351628 | 0.10 |
ENSMUST00000202365.2
ENSMUST00000186838.2 |
Gm42906
D730045B01Rik
|
predicted gene 42906 RIKEN cDNA D730045B01 gene |
| chr9_+_54493784 | 0.10 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr11_+_70416185 | 0.10 |
ENSMUST00000018430.7
|
Psmb6
|
proteasome (prosome, macropain) subunit, beta type 6 |
| chr5_-_35259847 | 0.10 |
ENSMUST00000153664.2
|
Lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr3_-_88204145 | 0.10 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr13_+_19398273 | 0.10 |
ENSMUST00000103558.3
|
Trgc1
|
T cell receptor gamma, constant 1 |
| chrX_-_94701983 | 0.10 |
ENSMUST00000119640.8
ENSMUST00000120620.8 ENSMUST00000044382.7 |
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr4_-_118294521 | 0.10 |
ENSMUST00000006565.13
|
Cdc20
|
cell division cycle 20 |
| chr8_+_47246534 | 0.10 |
ENSMUST00000210218.2
|
Irf2
|
interferon regulatory factor 2 |
| chr2_+_178056302 | 0.10 |
ENSMUST00000094251.11
|
Fam217b
|
family with sequence similarity 217, member B |
| chr5_-_23821523 | 0.10 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr11_+_80701001 | 0.10 |
ENSMUST00000040865.9
|
Tmem98
|
transmembrane protein 98 |
| chr3_-_36667610 | 0.10 |
ENSMUST00000108156.9
|
Bbs7
|
Bardet-Biedl syndrome 7 (human) |
| chr19_+_4242064 | 0.10 |
ENSMUST00000046094.6
|
Ppp1ca
|
protein phosphatase 1 catalytic subunit alpha |
| chr9_+_74959259 | 0.10 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_+_100900278 | 0.10 |
ENSMUST00000103110.10
ENSMUST00000044721.13 ENSMUST00000168757.9 |
Atp6v0a1
|
ATPase, H+ transporting, lysosomal V0 subunit A1 |
| chr8_+_95722289 | 0.10 |
ENSMUST00000211984.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr11_-_72106418 | 0.09 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr15_+_78294154 | 0.09 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr5_-_137500660 | 0.09 |
ENSMUST00000111035.2
ENSMUST00000031728.5 |
Pop7
|
processing of precursor 7, ribonuclease P family, (S. cerevisiae) |
| chr10_+_58207229 | 0.09 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr11_+_116089678 | 0.09 |
ENSMUST00000021130.7
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr2_+_87404246 | 0.09 |
ENSMUST00000213315.2
ENSMUST00000214773.2 |
Olfr1129
|
olfactory receptor 1129 |
| chr4_+_135648041 | 0.09 |
ENSMUST00000030434.5
|
Fuca1
|
fucosidase, alpha-L- 1, tissue |
| chr3_+_121746862 | 0.09 |
ENSMUST00000037958.14
ENSMUST00000196904.5 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr5_-_100521343 | 0.09 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr6_+_124807176 | 0.09 |
ENSMUST00000131847.8
ENSMUST00000151674.3 |
Cdca3
|
cell division cycle associated 3 |
| chr11_-_70130620 | 0.09 |
ENSMUST00000040428.4
|
Rnasek
|
ribonuclease, RNase K |
| chr1_+_88139678 | 0.09 |
ENSMUST00000073049.7
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr15_-_98575332 | 0.09 |
ENSMUST00000120997.2
ENSMUST00000109149.9 ENSMUST00000003451.11 |
Rnd1
|
Rho family GTPase 1 |
| chr5_-_69748126 | 0.09 |
ENSMUST00000166298.8
|
Gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr4_+_131570770 | 0.09 |
ENSMUST00000030742.11
ENSMUST00000137321.3 |
Mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr15_-_35938328 | 0.09 |
ENSMUST00000014457.15
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr7_+_143383363 | 0.09 |
ENSMUST00000124340.8
|
Dhcr7
|
7-dehydrocholesterol reductase |
| chr7_-_27749453 | 0.09 |
ENSMUST00000140053.3
ENSMUST00000032824.10 |
Psmc4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr12_-_46865709 | 0.09 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr6_+_124806506 | 0.09 |
ENSMUST00000150120.8
|
Cdca3
|
cell division cycle associated 3 |
| chr16_-_10360893 | 0.09 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr6_+_124806541 | 0.09 |
ENSMUST00000024270.14
|
Cdca3
|
cell division cycle associated 3 |
| chr10_+_93289264 | 0.08 |
ENSMUST00000016033.9
|
Lta4h
|
leukotriene A4 hydrolase |
| chr9_-_70328816 | 0.08 |
ENSMUST00000034742.8
|
Ccnb2
|
cyclin B2 |
| chr5_+_24305577 | 0.08 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr12_+_84408742 | 0.08 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr18_+_35904541 | 0.08 |
ENSMUST00000170693.9
ENSMUST00000237984.2 ENSMUST00000167406.2 |
Ube2d2a
|
ubiquitin-conjugating enzyme E2D 2A |
| chr9_+_57411279 | 0.08 |
ENSMUST00000080514.9
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr13_-_19491721 | 0.08 |
ENSMUST00000103561.3
|
Trgc2
|
T cell receptor gamma, constant 2 |
| chr1_+_88128323 | 0.08 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr17_+_79922329 | 0.08 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr11_+_78219241 | 0.08 |
ENSMUST00000048073.9
|
Pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr9_-_106769131 | 0.08 |
ENSMUST00000159283.8
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr2_-_155434487 | 0.08 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr4_+_155887978 | 0.08 |
ENSMUST00000030942.13
ENSMUST00000185148.8 ENSMUST00000130188.2 |
Mrpl20
|
mitochondrial ribosomal protein L20 |
| chr14_-_14651778 | 0.08 |
ENSMUST00000052678.9
|
Flnb
|
filamin, beta |
| chr8_-_4829519 | 0.08 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr9_+_64080644 | 0.08 |
ENSMUST00000034966.9
|
Rpl4
|
ribosomal protein L4 |
| chr14_+_70337540 | 0.08 |
ENSMUST00000022680.9
|
Bin3
|
bridging integrator 3 |
| chr10_+_88036947 | 0.08 |
ENSMUST00000020248.16
ENSMUST00000182183.8 ENSMUST00000171151.9 ENSMUST00000182619.2 |
Washc3
|
WASH complex subunit 3 |
| chr14_+_48683797 | 0.08 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr17_+_37269468 | 0.08 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr2_-_181333597 | 0.08 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr17_+_28396370 | 0.08 |
ENSMUST00000002318.8
ENSMUST00000233904.2 |
Zfp523
|
zinc finger protein 523 |
| chr19_-_46327071 | 0.08 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr8_-_41494890 | 0.08 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chrX_+_141009756 | 0.08 |
ENSMUST00000112916.9
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr18_+_3383230 | 0.08 |
ENSMUST00000162301.8
ENSMUST00000161317.2 |
Cul2
|
cullin 2 |
| chr16_+_38405718 | 0.08 |
ENSMUST00000165631.2
|
Tmem39a
|
transmembrane protein 39a |
| chr3_+_116306719 | 0.08 |
ENSMUST00000000349.11
ENSMUST00000197201.5 |
Dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr2_+_103788321 | 0.08 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr6_+_137731526 | 0.07 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr12_-_113802603 | 0.07 |
ENSMUST00000103458.3
ENSMUST00000193652.2 |
Ighv5-16
|
immunoglobulin heavy variable 5-16 |
| chr1_-_88437617 | 0.07 |
ENSMUST00000067625.9
|
Glrp1
|
glutamine repeat protein 1 |
| chr3_-_36667434 | 0.07 |
ENSMUST00000108155.8
|
Bbs7
|
Bardet-Biedl syndrome 7 (human) |
| chr5_-_130031681 | 0.07 |
ENSMUST00000111308.8
ENSMUST00000111307.2 |
Gusb
|
glucuronidase, beta |
| chrX_+_158086253 | 0.07 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr9_-_105398346 | 0.07 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr16_-_65359406 | 0.07 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr16_+_38383182 | 0.07 |
ENSMUST00000163948.8
|
Tmem39a
|
transmembrane protein 39a |
| chr1_-_24626492 | 0.07 |
ENSMUST00000051344.6
ENSMUST00000115244.9 |
Col19a1
|
collagen, type XIX, alpha 1 |
| chr19_-_44124252 | 0.07 |
ENSMUST00000026218.7
|
Cwf19l1
|
CWF19-like 1, cell cycle control (S. pombe) |
| chr15_-_55770118 | 0.07 |
ENSMUST00000110200.3
|
Sntb1
|
syntrophin, basic 1 |
| chr15_-_35938155 | 0.07 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr9_-_71803354 | 0.07 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr7_+_25380195 | 0.07 |
ENSMUST00000205658.2
|
B9d2
|
B9 protein domain 2 |
| chr7_+_25380263 | 0.07 |
ENSMUST00000205325.2
ENSMUST00000206913.2 |
B9d2
|
B9 protein domain 2 |
| chr14_-_52553764 | 0.07 |
ENSMUST00000135523.5
|
Sall2
|
spalt like transcription factor 2 |
| chr9_+_123901979 | 0.07 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chrX_+_133486391 | 0.07 |
ENSMUST00000113211.8
|
Rpl36a
|
ribosomal protein L36A |
| chr16_+_49519561 | 0.07 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr19_+_4282487 | 0.07 |
ENSMUST00000235306.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr18_-_67378886 | 0.07 |
ENSMUST00000073054.5
|
Mppe1
|
metallophosphoesterase 1 |
| chr4_+_3940747 | 0.07 |
ENSMUST00000119403.2
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_-_53771185 | 0.07 |
ENSMUST00000122330.2
ENSMUST00000146598.8 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chrX_-_20828242 | 0.07 |
ENSMUST00000123836.2
|
Uxt
|
ubiquitously expressed prefoldin like chaperone |
| chr14_+_46998004 | 0.07 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr1_+_172302925 | 0.07 |
ENSMUST00000027830.5
|
Slamf9
|
SLAM family member 9 |
| chr19_-_4675631 | 0.07 |
ENSMUST00000225375.2
ENSMUST00000025823.6 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr5_-_5470267 | 0.07 |
ENSMUST00000167567.2
|
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_+_59738866 | 0.07 |
ENSMUST00000102695.4
|
Nt5m
|
5',3'-nucleotidase, mitochondrial |
| chrX_+_154045439 | 0.07 |
ENSMUST00000026324.10
|
Acot9
|
acyl-CoA thioesterase 9 |
| chr10_+_77885768 | 0.07 |
ENSMUST00000151242.8
|
Dnmt3l
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr13_+_19528728 | 0.07 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr4_+_130253925 | 0.07 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr17_+_37269513 | 0.07 |
ENSMUST00000173814.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr2_+_4886298 | 0.07 |
ENSMUST00000027973.14
|
Sephs1
|
selenophosphate synthetase 1 |
| chr3_-_65300000 | 0.07 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr10_-_126866682 | 0.07 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr3_-_14676309 | 0.07 |
ENSMUST00000185423.2
ENSMUST00000186870.7 ENSMUST00000185384.7 |
Rbis
|
ribosomal biogenesis factor |
| chr7_-_98887770 | 0.07 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr11_-_83193412 | 0.07 |
ENSMUST00000176374.2
|
Pex12
|
peroxisomal biogenesis factor 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.1 | 1.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.3 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.3 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.1 | 0.3 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.4 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.2 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.1 | 0.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.2 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.1 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.0 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.0 | 0.0 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.0 | 0.1 | GO:1902870 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.1 | GO:0010924 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.0 | 0.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.0 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.0 | 0.0 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0070163 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.0 | 0.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.0 | GO:0070487 | monocyte aggregation(GO:0070487) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.0 | GO:1900108 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0042128 | nitrate assimilation(GO:0042128) |
| 0.0 | 0.1 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.0 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.0 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.3 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.2 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0009918 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.0 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.0 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.0 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |