Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
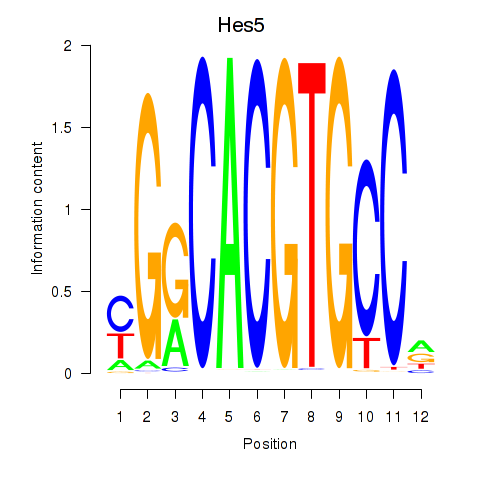
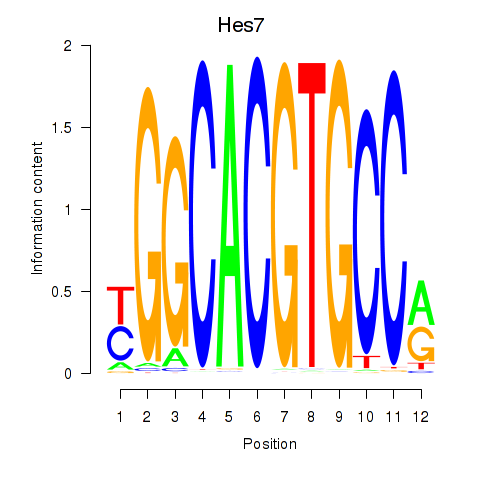
Results for Hes5_Hes7
Z-value: 0.50
Transcription factors associated with Hes5_Hes7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hes5
|
ENSMUSG00000048001.8 | hes family bHLH transcription factor 5 |
|
Hes7
|
ENSMUSG00000023781.3 | hes family bHLH transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hes5 | mm39_v1_chr4_+_155045372_155045387 | -0.76 | 1.4e-01 | Click! |
| Hes7 | mm39_v1_chr11_+_69011230_69011230 | -0.07 | 9.1e-01 | Click! |
Activity profile of Hes5_Hes7 motif
Sorted Z-values of Hes5_Hes7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_37004302 | 0.27 |
ENSMUST00000228275.2
|
Zfp706
|
zinc finger protein 706 |
| chr6_-_52203146 | 0.27 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr11_-_94932158 | 0.23 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr13_+_108452866 | 0.17 |
ENSMUST00000051594.12
|
Depdc1b
|
DEP domain containing 1B |
| chr12_+_77285770 | 0.17 |
ENSMUST00000062804.8
|
Fut8
|
fucosyltransferase 8 |
| chr11_+_54757200 | 0.16 |
ENSMUST00000020504.6
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr4_+_134195631 | 0.15 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr13_+_108452930 | 0.12 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr13_+_52000704 | 0.11 |
ENSMUST00000021903.3
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr2_+_150412329 | 0.10 |
ENSMUST00000089200.3
|
Cst7
|
cystatin F (leukocystatin) |
| chr14_-_33169099 | 0.10 |
ENSMUST00000111944.10
ENSMUST00000022504.12 ENSMUST00000111945.9 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr4_-_150998857 | 0.09 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr8_-_32499513 | 0.09 |
ENSMUST00000208205.3
|
Nrg1
|
neuregulin 1 |
| chr7_+_15795735 | 0.09 |
ENSMUST00000209369.2
|
Zfp541
|
zinc finger protein 541 |
| chr1_+_172327812 | 0.08 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr11_+_58197881 | 0.08 |
ENSMUST00000153510.9
|
Zfp692
|
zinc finger protein 692 |
| chr5_-_99876898 | 0.08 |
ENSMUST00000146396.8
ENSMUST00000161148.2 ENSMUST00000161516.2 |
Rasgef1b
|
RasGEF domain family, member 1B |
| chr10_-_80223475 | 0.08 |
ENSMUST00000105350.3
|
Mex3d
|
mex3 RNA binding family member D |
| chr1_-_75195889 | 0.08 |
ENSMUST00000186213.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr2_+_144441817 | 0.07 |
ENSMUST00000028917.7
|
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr17_-_25493273 | 0.07 |
ENSMUST00000172587.8
ENSMUST00000049911.16 ENSMUST00000173713.8 |
Ube2i
|
ubiquitin-conjugating enzyme E2I |
| chr11_-_50216426 | 0.07 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr13_-_10410857 | 0.07 |
ENSMUST00000187510.7
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr5_-_120610828 | 0.06 |
ENSMUST00000052258.14
ENSMUST00000031594.13 |
Sdsl
|
serine dehydratase-like |
| chr17_+_48623157 | 0.06 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr10_-_57408585 | 0.06 |
ENSMUST00000020027.11
|
Serinc1
|
serine incorporator 1 |
| chr11_-_54924231 | 0.05 |
ENSMUST00000108883.10
ENSMUST00000102727.3 |
Anxa6
|
annexin A6 |
| chr15_+_38662158 | 0.05 |
ENSMUST00000022904.8
ENSMUST00000228820.2 |
Atp6v1c1
|
ATPase, H+ transporting, lysosomal V1 subunit C1 |
| chr1_-_75195127 | 0.05 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr10_+_79518002 | 0.05 |
ENSMUST00000020550.13
|
Cdc34
|
cell division cycle 34 |
| chr6_+_49296208 | 0.05 |
ENSMUST00000055559.8
ENSMUST00000114491.2 |
Ccdc126
|
coiled-coil domain containing 126 |
| chr5_+_28370687 | 0.05 |
ENSMUST00000036177.9
|
En2
|
engrailed 2 |
| chr6_+_35154545 | 0.05 |
ENSMUST00000170234.2
|
Nup205
|
nucleoporin 205 |
| chr9_+_75221415 | 0.05 |
ENSMUST00000215875.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr9_+_55056648 | 0.05 |
ENSMUST00000121677.8
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr1_+_58432629 | 0.04 |
ENSMUST00000186949.2
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr4_+_155493668 | 0.04 |
ENSMUST00000123952.9
ENSMUST00000238423.2 ENSMUST00000238620.2 ENSMUST00000151083.8 |
Cfap74
|
cilia and flagella associated protein 74 |
| chr8_-_125448695 | 0.04 |
ENSMUST00000117624.9
ENSMUST00000231984.2 ENSMUST00000041614.15 ENSMUST00000214828.2 ENSMUST00000118134.3 |
Ttc13
|
tetratricopeptide repeat domain 13 |
| chr4_+_123798690 | 0.04 |
ENSMUST00000106202.4
|
Mycbp
|
MYC binding protein |
| chr17_-_24428351 | 0.04 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr6_-_145021816 | 0.04 |
ENSMUST00000111742.8
ENSMUST00000048252.11 |
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr10_-_87329513 | 0.04 |
ENSMUST00000020243.10
|
Ascl1
|
achaete-scute family bHLH transcription factor 1 |
| chr9_-_107109108 | 0.04 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr11_-_86698484 | 0.03 |
ENSMUST00000018569.14
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr11_+_100751272 | 0.03 |
ENSMUST00000107357.4
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr15_-_78739717 | 0.03 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr11_+_3913970 | 0.03 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr4_+_123798625 | 0.03 |
ENSMUST00000030400.14
|
Mycbp
|
MYC binding protein |
| chr6_+_71520855 | 0.03 |
ENSMUST00000204535.2
ENSMUST00000065364.5 ENSMUST00000204199.2 |
Chmp3
|
charged multivesicular body protein 3 |
| chr15_-_89258012 | 0.03 |
ENSMUST00000167643.4
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr19_-_5444413 | 0.03 |
ENSMUST00000044527.5
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr11_+_22462088 | 0.03 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr8_+_122996308 | 0.03 |
ENSMUST00000055537.3
|
Zfp469
|
zinc finger protein 469 |
| chr9_+_107464841 | 0.03 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr11_+_95557783 | 0.03 |
ENSMUST00000125172.8
ENSMUST00000036374.6 |
Phb
|
prohibitin |
| chr11_+_101207021 | 0.03 |
ENSMUST00000142640.8
ENSMUST00000019470.14 |
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chrX_-_8042129 | 0.03 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr9_+_64028783 | 0.02 |
ENSMUST00000118215.3
|
Lctl
|
lactase-like |
| chr17_-_15181491 | 0.02 |
ENSMUST00000024657.12
|
Phf10
|
PHD finger protein 10 |
| chr7_-_130964469 | 0.02 |
ENSMUST00000059438.11
|
2310057M21Rik
|
RIKEN cDNA 2310057M21 gene |
| chr10_+_79518138 | 0.02 |
ENSMUST00000166603.2
ENSMUST00000219791.2 ENSMUST00000219930.2 ENSMUST00000218964.2 |
Cdc34
|
cell division cycle 34 |
| chr11_-_11976732 | 0.02 |
ENSMUST00000143915.2
|
Grb10
|
growth factor receptor bound protein 10 |
| chr11_-_11976237 | 0.02 |
ENSMUST00000150972.8
|
Grb10
|
growth factor receptor bound protein 10 |
| chr4_-_138484889 | 0.02 |
ENSMUST00000030526.7
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr17_+_36172210 | 0.02 |
ENSMUST00000074259.15
ENSMUST00000174873.2 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr9_-_121470564 | 0.02 |
ENSMUST00000213757.2
ENSMUST00000125075.2 ENSMUST00000077706.10 ENSMUST00000214592.2 |
Lyzl4
|
lysozyme-like 4 |
| chr8_-_84769170 | 0.02 |
ENSMUST00000005601.9
|
Il27ra
|
interleukin 27 receptor, alpha |
| chr1_-_192718064 | 0.02 |
ENSMUST00000215093.2
ENSMUST00000195354.6 |
Syt14
|
synaptotagmin XIV |
| chr13_+_43938251 | 0.02 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr18_+_37276556 | 0.01 |
ENSMUST00000047479.3
|
Pcdhac2
|
protocadherin alpha subfamily C, 2 |
| chr4_-_117109074 | 0.01 |
ENSMUST00000165128.9
|
Armh1
|
armadillo-like helical domain containing 1 |
| chr10_+_97528915 | 0.01 |
ENSMUST00000060703.6
|
Ccer1
|
coiled-coil glutamate-rich protein 1 |
| chr5_+_122788469 | 0.01 |
ENSMUST00000199371.2
|
P2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr10_-_62486948 | 0.01 |
ENSMUST00000020270.6
|
Ddx50
|
DExD box helicase 50 |
| chr13_+_49806772 | 0.01 |
ENSMUST00000223264.2
ENSMUST00000221142.2 ENSMUST00000222333.2 ENSMUST00000021824.8 |
Nol8
|
nucleolar protein 8 |
| chr9_-_39684094 | 0.01 |
ENSMUST00000075928.6
|
Olfr968
|
olfactory receptor 968 |
| chr11_+_79883885 | 0.01 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr8_+_120173458 | 0.01 |
ENSMUST00000098363.10
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr4_-_132484559 | 0.01 |
ENSMUST00000030709.9
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr19_+_47167259 | 0.01 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr2_-_65397850 | 0.01 |
ENSMUST00000238483.2
ENSMUST00000100069.9 |
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr2_+_91357100 | 0.01 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr4_-_43562397 | 0.01 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr3_+_95466982 | 0.01 |
ENSMUST00000090797.11
ENSMUST00000171191.6 ENSMUST00000029754.13 ENSMUST00000107154.4 |
Hormad1
|
HORMA domain containing 1 |
| chr1_+_166081664 | 0.01 |
ENSMUST00000111416.7
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr11_+_59197746 | 0.01 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr16_-_33787399 | 0.01 |
ENSMUST00000023510.7
|
Umps
|
uridine monophosphate synthetase |
| chr14_-_69945022 | 0.01 |
ENSMUST00000118374.8
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr6_+_72074545 | 0.01 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr5_+_43672856 | 0.01 |
ENSMUST00000076939.10
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr3_-_95789505 | 0.01 |
ENSMUST00000159863.2
ENSMUST00000159739.8 ENSMUST00000036418.10 ENSMUST00000161866.8 |
Ciart
|
circadian associated repressor of transcription |
| chr9_-_35469818 | 0.01 |
ENSMUST00000034612.7
|
Ddx25
|
DEAD box helicase 25 |
| chr15_+_81954197 | 0.01 |
ENSMUST00000229119.2
ENSMUST00000089178.11 |
Mei1
|
meiotic double-stranded break formation protein 1 |
| chr11_+_103540391 | 0.01 |
ENSMUST00000057870.4
|
Rprml
|
reprimo-like |
| chr1_+_166081755 | 0.01 |
ENSMUST00000194964.6
ENSMUST00000192638.6 ENSMUST00000192426.6 ENSMUST00000195557.6 ENSMUST00000192732.6 ENSMUST00000193860.2 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr7_+_119927885 | 0.01 |
ENSMUST00000207220.2
ENSMUST00000121265.7 ENSMUST00000076272.5 |
Abca15
|
ATP-binding cassette, sub-family A (ABC1), member 15 |
| chr12_-_72964534 | 0.01 |
ENSMUST00000110489.9
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr12_+_72807985 | 0.01 |
ENSMUST00000021514.10
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform |
| chr11_+_54757239 | 0.01 |
ENSMUST00000117710.2
|
Hint1
|
histidine triad nucleotide binding protein 1 |
| chr6_+_64706101 | 0.01 |
ENSMUST00000101351.6
|
Atoh1
|
atonal bHLH transcription factor 1 |
| chr17_-_3746536 | 0.01 |
ENSMUST00000115800.2
|
Nox3
|
NADPH oxidase 3 |
| chr13_+_98439012 | 0.00 |
ENSMUST00000186911.2
|
Gm21976
|
predicted gene 21976 |
| chr12_+_16703383 | 0.00 |
ENSMUST00000221596.2
ENSMUST00000111064.3 ENSMUST00000220892.2 |
Ntsr2
|
neurotensin receptor 2 |
| chr5_+_92534738 | 0.00 |
ENSMUST00000128246.8
|
Art3
|
ADP-ribosyltransferase 3 |
| chr15_-_95426108 | 0.00 |
ENSMUST00000075275.3
|
Nell2
|
NEL-like 2 |
| chr3_-_95789338 | 0.00 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_65397809 | 0.00 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr19_+_8713156 | 0.00 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr1_-_192717958 | 0.00 |
ENSMUST00000016344.9
|
Syt14
|
synaptotagmin XIV |
| chr5_+_43673093 | 0.00 |
ENSMUST00000144558.3
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr4_+_137640782 | 0.00 |
ENSMUST00000151110.4
|
Ece1
|
endothelin converting enzyme 1 |
| chr7_-_142522454 | 0.00 |
ENSMUST00000121862.3
|
Ascl2
|
achaete-scute family bHLH transcription factor 2 |
| chr12_+_16703709 | 0.00 |
ENSMUST00000221049.2
|
Ntsr2
|
neurotensin receptor 2 |
| chr13_-_22363752 | 0.00 |
ENSMUST00000072369.2
|
Vmn1r191
|
vomeronasal 1 receptor 191 |
| chr8_-_25438784 | 0.00 |
ENSMUST00000119720.8
ENSMUST00000121438.9 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr1_+_92500847 | 0.00 |
ENSMUST00000074859.3
|
Olfr1413
|
olfactory receptor 1413 |
| chr5_-_135773047 | 0.00 |
ENSMUST00000153399.2
|
Tmem120a
|
transmembrane protein 120A |
| chr8_-_88531018 | 0.00 |
ENSMUST00000165770.9
|
Zfp423
|
zinc finger protein 423 |
| chr13_-_32960379 | 0.00 |
ENSMUST00000230119.2
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr1_+_192856044 | 0.00 |
ENSMUST00000193307.2
|
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr18_+_11972277 | 0.00 |
ENSMUST00000171109.9
ENSMUST00000046948.10 |
Cables1
|
CDK5 and Abl enzyme substrate 1 |
| chr19_+_46120327 | 0.00 |
ENSMUST00000043739.6
ENSMUST00000237098.2 |
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr2_-_92201311 | 0.00 |
ENSMUST00000176774.8
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr2_-_155534295 | 0.00 |
ENSMUST00000041059.12
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr13_+_49806542 | 0.00 |
ENSMUST00000222197.2
ENSMUST00000221083.2 ENSMUST00000223467.2 |
Nol8
|
nucleolar protein 8 |
| chr5_+_128677863 | 0.00 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr10_+_85665234 | 0.00 |
ENSMUST00000217667.2
|
Pwp1
|
PWP1 homolog, endonuclein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hes5_Hes7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:1903189 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) olfactory pit development(GO:0060166) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.0 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |