Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
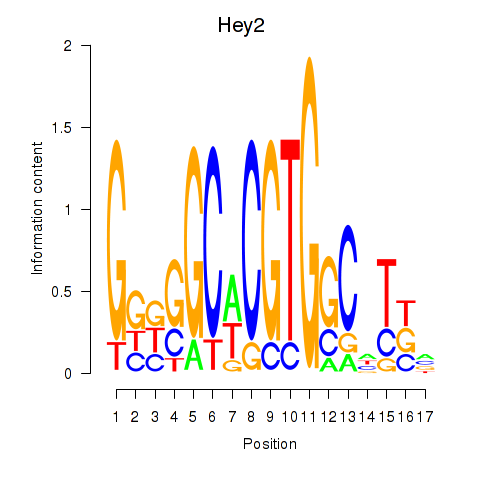
Results for Hey2
Z-value: 1.34
Transcription factors associated with Hey2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hey2
|
ENSMUSG00000019789.10 | hairy/enhancer-of-split related with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hey2 | mm39_v1_chr10_-_30718760_30718797 | -0.60 | 2.8e-01 | Click! |
Activity profile of Hey2 motif
Sorted Z-values of Hey2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_30741012 | 1.29 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr10_+_128745214 | 0.99 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr14_-_30740946 | 0.94 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chrX_+_68403900 | 0.69 |
ENSMUST00000033532.7
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr11_+_103857541 | 0.69 |
ENSMUST00000057921.10
ENSMUST00000063347.12 |
Arf2
|
ADP-ribosylation factor 2 |
| chr7_+_100970435 | 0.68 |
ENSMUST00000210192.2
ENSMUST00000172630.8 |
Stard10
|
START domain containing 10 |
| chr14_-_31552335 | 0.65 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr2_+_127112127 | 0.61 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr5_+_52898910 | 0.59 |
ENSMUST00000031081.11
ENSMUST00000031082.8 |
Pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr11_-_93846453 | 0.59 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr18_+_68433422 | 0.57 |
ENSMUST00000009679.11
ENSMUST00000131075.8 ENSMUST00000025427.14 ENSMUST00000139111.2 |
Rnmt
|
RNA (guanine-7-) methyltransferase |
| chr4_+_134195631 | 0.55 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr4_-_120427449 | 0.54 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chr10_+_61431271 | 0.52 |
ENSMUST00000020287.8
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr7_+_100970910 | 0.49 |
ENSMUST00000174291.8
ENSMUST00000167888.9 ENSMUST00000172662.2 |
Stard10
|
START domain containing 10 |
| chr3_-_88410495 | 0.48 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr10_+_127919142 | 0.47 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr10_-_112764879 | 0.47 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr9_+_22068197 | 0.46 |
ENSMUST00000148088.2
|
Pigyl
|
phosphatidylinositol glycan anchor biosynthesis, class Y-like |
| chr12_+_117807224 | 0.45 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like |
| chr1_+_180731843 | 0.44 |
ENSMUST00000027802.9
|
Pycr2
|
pyrroline-5-carboxylate reductase family, member 2 |
| chr7_-_90106375 | 0.44 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A |
| chr3_+_104545974 | 0.44 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr1_-_30988772 | 0.43 |
ENSMUST00000238874.2
ENSMUST00000027232.15 ENSMUST00000076587.6 ENSMUST00000233506.2 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr5_-_44383943 | 0.43 |
ENSMUST00000055128.12
|
Tapt1
|
transmembrane anterior posterior transformation 1 |
| chr19_+_6952580 | 0.42 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr7_-_125968653 | 0.42 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr9_+_7184514 | 0.42 |
ENSMUST00000215683.2
ENSMUST00000034499.10 |
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr7_-_64041996 | 0.42 |
ENSMUST00000032735.8
|
Mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr8_-_111629074 | 0.40 |
ENSMUST00000041382.7
|
Fcsk
|
fucose kinase |
| chr9_+_66620959 | 0.40 |
ENSMUST00000071889.13
|
Car12
|
carbonic anhydrase 12 |
| chr1_-_106687457 | 0.40 |
ENSMUST00000010049.6
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr5_-_122639840 | 0.40 |
ENSMUST00000177974.8
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr11_-_58059293 | 0.39 |
ENSMUST00000172035.8
ENSMUST00000035604.13 ENSMUST00000102711.9 |
Gemin5
|
gem nuclear organelle associated protein 5 |
| chr19_+_6952319 | 0.39 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr1_-_30988381 | 0.39 |
ENSMUST00000232841.2
|
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr9_+_66621001 | 0.38 |
ENSMUST00000085420.12
|
Car12
|
carbonic anhydrase 12 |
| chr17_-_25105277 | 0.38 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr12_+_76353835 | 0.38 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr17_-_31731222 | 0.36 |
ENSMUST00000236665.2
|
Wdr4
|
WD repeat domain 4 |
| chr3_+_103875261 | 0.36 |
ENSMUST00000117150.8
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr10_+_7543260 | 0.35 |
ENSMUST00000040135.9
|
Nup43
|
nucleoporin 43 |
| chr3_+_103875574 | 0.35 |
ENSMUST00000063717.14
ENSMUST00000055425.15 ENSMUST00000123611.8 ENSMUST00000090685.11 |
Phtf1
|
putative homeodomain transcription factor 1 |
| chr13_-_94422337 | 0.34 |
ENSMUST00000022197.15
ENSMUST00000152555.8 |
Scamp1
|
secretory carrier membrane protein 1 |
| chr8_+_80366247 | 0.33 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chr4_+_28813125 | 0.33 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr4_-_11965691 | 0.32 |
ENSMUST00000108301.8
ENSMUST00000095144.10 ENSMUST00000108302.8 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr7_+_97345841 | 0.31 |
ENSMUST00000026506.5
|
Clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr2_+_127112613 | 0.31 |
ENSMUST00000125049.2
ENSMUST00000110374.2 |
Stard7
|
START domain containing 7 |
| chr14_-_70588803 | 0.30 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr13_-_104246084 | 0.30 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr5_+_143534455 | 0.30 |
ENSMUST00000169329.8
ENSMUST00000067145.12 ENSMUST00000119488.2 ENSMUST00000118121.2 ENSMUST00000200267.2 ENSMUST00000196487.2 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr4_+_28813152 | 0.30 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chrX_+_49930311 | 0.29 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr2_+_122065230 | 0.29 |
ENSMUST00000110551.4
|
Sord
|
sorbitol dehydrogenase |
| chr19_+_36811615 | 0.27 |
ENSMUST00000025729.12
|
Tnks2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr9_+_107464841 | 0.27 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr3_-_89325594 | 0.27 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr12_+_117807607 | 0.26 |
ENSMUST00000176735.8
ENSMUST00000177339.2 |
Cdca7l
|
cell division cycle associated 7 like |
| chr5_-_88823049 | 0.26 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr9_-_53521585 | 0.26 |
ENSMUST00000034547.6
|
Acat1
|
acetyl-Coenzyme A acetyltransferase 1 |
| chr3_+_40904253 | 0.26 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr8_-_71834543 | 0.26 |
ENSMUST00000002466.9
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr5_+_139777263 | 0.25 |
ENSMUST00000018287.10
|
Mafk
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
| chr7_-_51511964 | 0.25 |
ENSMUST00000169357.2
|
Fancf
|
Fanconi anemia, complementation group F |
| chr7_-_30810422 | 0.25 |
ENSMUST00000039435.15
|
Hpn
|
hepsin |
| chr10_+_79977291 | 0.24 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr5_+_91079068 | 0.24 |
ENSMUST00000202781.2
ENSMUST00000071652.6 |
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr5_-_121523670 | 0.23 |
ENSMUST00000146185.2
ENSMUST00000042312.14 |
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr4_+_116734573 | 0.23 |
ENSMUST00000044823.4
|
Zswim5
|
zinc finger SWIM-type containing 5 |
| chr5_+_100993996 | 0.23 |
ENSMUST00000112887.8
ENSMUST00000031255.15 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr18_-_36587573 | 0.23 |
ENSMUST00000025204.7
ENSMUST00000237792.2 |
Pfdn1
|
prefoldin 1 |
| chr10_-_62486948 | 0.22 |
ENSMUST00000020270.6
|
Ddx50
|
DExD box helicase 50 |
| chr1_-_93659622 | 0.22 |
ENSMUST00000189728.7
|
Thap4
|
THAP domain containing 4 |
| chr8_-_94739469 | 0.21 |
ENSMUST00000053766.14
|
Amfr
|
autocrine motility factor receptor |
| chr9_+_22068134 | 0.20 |
ENSMUST00000123680.2
|
Pigyl
|
phosphatidylinositol glycan anchor biosynthesis, class Y-like |
| chr8_-_23747023 | 0.20 |
ENSMUST00000121783.7
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr15_+_58761057 | 0.20 |
ENSMUST00000036904.7
|
Rnf139
|
ring finger protein 139 |
| chr8_+_85786684 | 0.19 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr3_-_90297187 | 0.19 |
ENSMUST00000029541.12
|
Slc27a3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chrX_+_70408351 | 0.19 |
ENSMUST00000146213.8
ENSMUST00000114601.8 ENSMUST00000015358.8 |
Mtmr1
|
myotubularin related protein 1 |
| chr19_-_55087849 | 0.19 |
ENSMUST00000061856.6
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr6_+_125108829 | 0.18 |
ENSMUST00000044200.11
ENSMUST00000204185.2 |
Nop2
|
NOP2 nucleolar protein |
| chr13_-_100969823 | 0.18 |
ENSMUST00000225922.2
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr9_+_48896765 | 0.18 |
ENSMUST00000047349.8
|
Usp28
|
ubiquitin specific peptidase 28 |
| chrX_-_7940959 | 0.17 |
ENSMUST00000115636.4
ENSMUST00000115638.10 |
Suv39h1
|
suppressor of variegation 3-9 1 |
| chr17_+_73144531 | 0.17 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr16_+_91855158 | 0.17 |
ENSMUST00000047429.9
ENSMUST00000232677.2 ENSMUST00000113975.3 |
Mrps6
Gm49711
Slc5a3
|
mitochondrial ribosomal protein S6 predicted gene, 49711 solute carrier family 5 (inositol transporters), member 3 |
| chr7_+_100971034 | 0.17 |
ENSMUST00000173270.8
|
Stard10
|
START domain containing 10 |
| chr3_+_95222102 | 0.17 |
ENSMUST00000129267.8
|
Cers2
|
ceramide synthase 2 |
| chr1_+_59724108 | 0.16 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr2_+_29956432 | 0.16 |
ENSMUST00000067996.7
|
Set
|
SET nuclear oncogene |
| chr17_-_31731190 | 0.16 |
ENSMUST00000237127.2
|
Wdr4
|
WD repeat domain 4 |
| chr18_+_67422256 | 0.16 |
ENSMUST00000025403.8
|
Impa2
|
inositol monophosphatase 2 |
| chr13_+_51562675 | 0.15 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr8_-_71060911 | 0.15 |
ENSMUST00000210580.2
ENSMUST00000211608.2 ENSMUST00000049908.11 |
Ssbp4
|
single stranded DNA binding protein 4 |
| chr15_+_38976260 | 0.15 |
ENSMUST00000022909.10
|
Dcaf13
|
DDB1 and CUL4 associated factor 13 |
| chr9_-_36637670 | 0.14 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr9_+_60701749 | 0.14 |
ENSMUST00000214354.2
ENSMUST00000050183.7 ENSMUST00000217656.2 |
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr13_-_100969878 | 0.14 |
ENSMUST00000067246.6
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr11_+_97697328 | 0.13 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr11_+_102080489 | 0.13 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr3_+_81839908 | 0.13 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chrX_+_70408507 | 0.13 |
ENSMUST00000132837.5
|
Mtmr1
|
myotubularin related protein 1 |
| chr5_-_122640255 | 0.13 |
ENSMUST00000031423.10
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr12_+_30961650 | 0.13 |
ENSMUST00000020997.15
ENSMUST00000110880.3 |
Sh3yl1
|
Sh3 domain YSC-like 1 |
| chr5_+_100994230 | 0.13 |
ENSMUST00000092990.4
ENSMUST00000145612.2 |
Gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr8_+_72021510 | 0.12 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr15_-_38976034 | 0.12 |
ENSMUST00000227323.2
ENSMUST00000022908.10 |
Slc25a32
|
solute carrier family 25, member 32 |
| chr1_+_164076592 | 0.12 |
ENSMUST00000044021.12
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr9_-_36637923 | 0.12 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr1_-_120192977 | 0.12 |
ENSMUST00000140490.8
ENSMUST00000112640.8 |
Steap3
|
STEAP family member 3 |
| chr12_-_27392356 | 0.12 |
ENSMUST00000079063.7
|
Sox11
|
SRY (sex determining region Y)-box 11 |
| chr17_-_46342739 | 0.11 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr10_+_80971054 | 0.11 |
ENSMUST00000125261.2
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr11_+_72498029 | 0.11 |
ENSMUST00000021148.13
ENSMUST00000138247.8 |
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr8_+_72021567 | 0.11 |
ENSMUST00000034267.5
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr12_+_86725459 | 0.10 |
ENSMUST00000021681.4
|
Vash1
|
vasohibin 1 |
| chr1_+_164076621 | 0.10 |
ENSMUST00000159230.8
|
Slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr11_+_102080446 | 0.09 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr6_-_88851579 | 0.09 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr15_-_79339727 | 0.09 |
ENSMUST00000230599.2
|
Csnk1e
|
casein kinase 1, epsilon |
| chr9_+_48896656 | 0.08 |
ENSMUST00000215788.2
ENSMUST00000213874.2 |
Usp28
|
ubiquitin specific peptidase 28 |
| chr7_+_79910948 | 0.08 |
ENSMUST00000117989.2
|
Ngrn
|
neugrin, neurite outgrowth associated |
| chr8_-_23747057 | 0.07 |
ENSMUST00000051094.9
|
Golga7
|
golgi autoantigen, golgin subfamily a, 7 |
| chr1_+_21288799 | 0.07 |
ENSMUST00000027065.12
|
Tmem14a
|
transmembrane protein 14A |
| chr4_-_148172423 | 0.07 |
ENSMUST00000030865.9
|
Agtrap
|
angiotensin II, type I receptor-associated protein |
| chr5_+_67418137 | 0.06 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr4_-_117013396 | 0.06 |
ENSMUST00000102696.5
|
Rps8
|
ribosomal protein S8 |
| chr1_-_37469037 | 0.06 |
ENSMUST00000027286.7
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr17_-_88105422 | 0.06 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr3_+_51131868 | 0.06 |
ENSMUST00000023849.15
ENSMUST00000167780.2 |
Noct
|
nocturnin |
| chr7_-_128342158 | 0.06 |
ENSMUST00000119081.2
ENSMUST00000057557.14 |
Mcmbp
|
minichromosome maintenance complex binding protein |
| chr8_+_4399588 | 0.05 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr9_-_114762986 | 0.05 |
ENSMUST00000146623.8
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr7_-_29935150 | 0.05 |
ENSMUST00000189482.2
|
Ovol3
|
ovo like zinc finger 3 |
| chrX_+_139565657 | 0.05 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr19_+_22116392 | 0.05 |
ENSMUST00000237357.2
ENSMUST00000237820.2 ENSMUST00000236312.2 ENSMUST00000074770.13 ENSMUST00000087576.12 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr9_-_114762879 | 0.05 |
ENSMUST00000084853.4
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr3_+_28835425 | 0.05 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr11_-_106278892 | 0.04 |
ENSMUST00000106813.9
ENSMUST00000141146.2 |
Icam2
|
intercellular adhesion molecule 2 |
| chr13_+_110531571 | 0.04 |
ENSMUST00000022212.9
|
Plk2
|
polo like kinase 2 |
| chr5_-_44259010 | 0.04 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr11_+_97690585 | 0.04 |
ENSMUST00000129558.8
|
Lasp1
|
LIM and SH3 protein 1 |
| chr15_+_97862081 | 0.04 |
ENSMUST00000229433.2
ENSMUST00000229428.2 ENSMUST00000064200.9 ENSMUST00000230072.2 ENSMUST00000231144.2 |
Tmem106c
|
transmembrane protein 106C |
| chr10_+_127575407 | 0.04 |
ENSMUST00000054287.9
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr4_-_126647156 | 0.04 |
ENSMUST00000030637.14
ENSMUST00000106116.2 |
Ncdn
|
neurochondrin |
| chr1_+_21288886 | 0.04 |
ENSMUST00000027064.8
|
Tmem14a
|
transmembrane protein 14A |
| chr18_-_68433398 | 0.03 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr11_+_97690391 | 0.03 |
ENSMUST00000043843.12
|
Lasp1
|
LIM and SH3 protein 1 |
| chr4_+_155334417 | 0.03 |
ENSMUST00000178473.8
ENSMUST00000105627.8 ENSMUST00000097747.9 |
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr14_-_71003973 | 0.03 |
ENSMUST00000226448.2
ENSMUST00000022696.8 |
Xpo7
|
exportin 7 |
| chr14_+_33041660 | 0.03 |
ENSMUST00000111955.2
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr9_-_103357564 | 0.03 |
ENSMUST00000124310.5
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr7_+_101916992 | 0.03 |
ENSMUST00000033289.6
ENSMUST00000209255.2 |
Stim1
|
stromal interaction molecule 1 |
| chr13_-_41373638 | 0.03 |
ENSMUST00000117096.2
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr5_-_8417982 | 0.02 |
ENSMUST00000088761.11
ENSMUST00000115386.8 ENSMUST00000050166.14 ENSMUST00000046838.14 ENSMUST00000115388.9 ENSMUST00000088744.12 ENSMUST00000115385.2 |
Adam22
|
a disintegrin and metallopeptidase domain 22 |
| chr11_-_99742434 | 0.02 |
ENSMUST00000107437.2
|
Krtap4-16
|
keratin associated protein 4-16 |
| chr13_-_114595475 | 0.02 |
ENSMUST00000022287.8
ENSMUST00000223640.3 |
Fst
|
follistatin |
| chr6_-_89339581 | 0.02 |
ENSMUST00000163139.8
|
Plxna1
|
plexin A1 |
| chr13_-_23012284 | 0.02 |
ENSMUST00000072044.2
|
Vmn1r210
|
vomeronasal 1 receptor 210 |
| chr11_+_118319319 | 0.02 |
ENSMUST00000017590.9
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr13_-_14697681 | 0.02 |
ENSMUST00000223189.2
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr16_-_4442802 | 0.02 |
ENSMUST00000014445.7
|
Pam16
|
presequence translocase-asssociated motor 16 homolog (S. cerevisiae) |
| chr18_-_68433265 | 0.02 |
ENSMUST00000152193.2
|
Fam210a
|
family with sequence similarity 210, member A |
| chr8_-_47742429 | 0.02 |
ENSMUST00000079195.6
|
Stox2
|
storkhead box 2 |
| chr9_+_39933648 | 0.02 |
ENSMUST00000059859.5
|
Olfr981
|
olfactory receptor 981 |
| chr2_+_31840151 | 0.02 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr10_-_117212860 | 0.01 |
ENSMUST00000069168.13
ENSMUST00000176686.8 |
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr10_-_10956700 | 0.01 |
ENSMUST00000105560.2
|
Grm1
|
glutamate receptor, metabotropic 1 |
| chr13_-_25454058 | 0.01 |
ENSMUST00000057866.13
|
Nrsn1
|
neurensin 1 |
| chr13_+_58955675 | 0.01 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr13_-_114595122 | 0.01 |
ENSMUST00000231252.2
|
Fst
|
follistatin |
| chr18_+_62457275 | 0.01 |
ENSMUST00000027560.8
|
Htr4
|
5 hydroxytryptamine (serotonin) receptor 4 |
| chr9_+_57604895 | 0.01 |
ENSMUST00000034865.6
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr4_-_108240546 | 0.01 |
ENSMUST00000053157.7
|
Shisal2a
|
shisa like 2A |
| chr14_-_55163452 | 0.01 |
ENSMUST00000227037.2
|
Efs
|
embryonal Fyn-associated substrate |
| chr19_-_6910922 | 0.01 |
ENSMUST00000235248.2
|
Kcnk4
|
potassium channel, subfamily K, member 4 |
| chr4_+_104224774 | 0.01 |
ENSMUST00000106830.9
|
Dab1
|
disabled 1 |
| chr2_-_113844100 | 0.01 |
ENSMUST00000090275.5
|
Gjd2
|
gap junction protein, delta 2 |
| chr2_+_31840340 | 0.01 |
ENSMUST00000148056.4
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr2_-_179746227 | 0.01 |
ENSMUST00000056480.10
|
Hrh3
|
histamine receptor H3 |
| chr6_+_17306379 | 0.00 |
ENSMUST00000115455.3
|
Cav1
|
caveolin 1, caveolae protein |
| chr17_-_33435325 | 0.00 |
ENSMUST00000112162.4
|
Olfr1564
|
olfactory receptor 1564 |
| chr1_+_120268299 | 0.00 |
ENSMUST00000037286.12
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr5_+_53206688 | 0.00 |
ENSMUST00000094787.8
|
Slc34a2
|
solute carrier family 34 (sodium phosphate), member 2 |
| chr4_+_101276893 | 0.00 |
ENSMUST00000131397.2
ENSMUST00000133055.8 |
Ak4
|
adenylate kinase 4 |
| chr15_-_7427759 | 0.00 |
ENSMUST00000058593.10
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr7_+_51160855 | 0.00 |
ENSMUST00000207717.2
|
Ano5
|
anoctamin 5 |
| chr5_-_122187884 | 0.00 |
ENSMUST00000111752.10
|
Cux2
|
cut-like homeobox 2 |
| chr13_-_14697770 | 0.00 |
ENSMUST00000110516.3
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_103209686 | 0.00 |
ENSMUST00000098198.3
|
Olfr615
|
olfactory receptor 615 |
| chr7_+_49624978 | 0.00 |
ENSMUST00000107603.2
|
Nell1
|
NEL-like 1 |
| chr15_-_7427815 | 0.00 |
ENSMUST00000096494.5
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr15_+_98687720 | 0.00 |
ENSMUST00000023734.8
|
Wnt1
|
wingless-type MMTV integration site family, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hey2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.2 | 1.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 0.6 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.1 | 1.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 2.2 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.3 | GO:0019405 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.1 | 0.3 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.1 | 0.3 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.8 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.1 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.4 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.4 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.1 | GO:1903896 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.2 | 0.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.6 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.5 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.6 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.2 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.6 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0052851 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |