Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
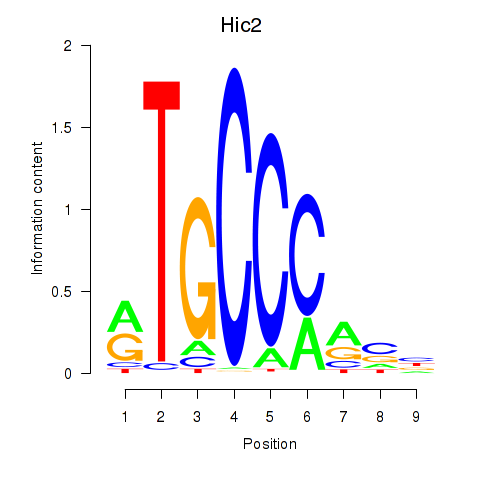
Results for Hic2
Z-value: 1.34
Transcription factors associated with Hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hic2
|
ENSMUSG00000050240.17 | hypermethylated in cancer 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic2 | mm39_v1_chr16_+_17051423_17051528 | -0.69 | 2.0e-01 | Click! |
Activity profile of Hic2 motif
Sorted Z-values of Hic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23758370 | 1.64 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr13_+_21938258 | 1.15 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr13_-_21994366 | 0.85 |
ENSMUST00000091749.4
|
H2bc23
|
H2B clustered histone 23 |
| chr13_-_21900313 | 0.75 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr17_+_7292967 | 0.61 |
ENSMUST00000097422.6
|
Gm1604b
|
predicted gene 1604b |
| chr8_-_69541852 | 0.54 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr9_+_118881838 | 0.49 |
ENSMUST00000051386.13
ENSMUST00000074734.13 |
Vill
|
villin-like |
| chr11_-_94932158 | 0.48 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr15_-_101268036 | 0.47 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr7_-_6525801 | 0.47 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr15_+_102368510 | 0.40 |
ENSMUST00000164688.2
|
Prr13
|
proline rich 13 |
| chr6_+_88175312 | 0.38 |
ENSMUST00000203480.2
ENSMUST00000015197.9 |
Gata2
|
GATA binding protein 2 |
| chr15_+_31224555 | 0.38 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr11_-_78313043 | 0.37 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr7_+_46700349 | 0.37 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A |
| chrX_-_72924436 | 0.36 |
ENSMUST00000102871.10
|
L1cam
|
L1 cell adhesion molecule |
| chr2_-_28945051 | 0.36 |
ENSMUST00000189711.7
ENSMUST00000157048.4 |
Cfap77
|
cilia and flagella associated protein 77 |
| chr7_+_142030744 | 0.36 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr14_-_55150547 | 0.35 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr7_-_30119227 | 0.35 |
ENSMUST00000208740.2
ENSMUST00000075062.5 |
Hcst
|
hematopoietic cell signal transducer |
| chr19_+_11382092 | 0.34 |
ENSMUST00000153546.8
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C |
| chr5_-_23881353 | 0.34 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr6_-_146403410 | 0.34 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr2_+_164790139 | 0.33 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr5_-_30619246 | 0.33 |
ENSMUST00000114747.9
ENSMUST00000074171.10 |
Otof
|
otoferlin |
| chr4_-_133856025 | 0.32 |
ENSMUST00000105879.2
ENSMUST00000030651.9 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr15_+_31224616 | 0.32 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr3_+_5815863 | 0.31 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr2_-_30305779 | 0.31 |
ENSMUST00000102855.8
ENSMUST00000028207.13 |
Crat
|
carnitine acetyltransferase |
| chr17_-_48474356 | 0.31 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr7_+_18786237 | 0.30 |
ENSMUST00000130328.2
|
Sympk
|
symplekin |
| chr4_+_133829898 | 0.29 |
ENSMUST00000070246.9
ENSMUST00000156750.2 |
Ubxn11
|
UBX domain protein 11 |
| chr6_-_149003171 | 0.29 |
ENSMUST00000111557.8
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr7_-_43956326 | 0.28 |
ENSMUST00000004587.11
|
Clec11a
|
C-type lectin domain family 11, member a |
| chr6_-_149003003 | 0.28 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chr11_+_115768323 | 0.28 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr4_-_140501507 | 0.27 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr6_-_146403638 | 0.27 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr17_+_31276649 | 0.27 |
ENSMUST00000236391.2
ENSMUST00000024829.8 ENSMUST00000236427.2 |
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr2_-_170269748 | 0.27 |
ENSMUST00000013667.3
ENSMUST00000109152.9 ENSMUST00000068137.11 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr6_-_52185674 | 0.26 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr7_-_24459736 | 0.26 |
ENSMUST00000063956.7
|
Cd177
|
CD177 antigen |
| chr17_+_48623157 | 0.26 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr2_+_152578164 | 0.26 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr8_+_13455080 | 0.25 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chr3_+_89084770 | 0.25 |
ENSMUST00000029684.15
ENSMUST00000120697.8 ENSMUST00000098941.5 |
Scamp3
|
secretory carrier membrane protein 3 |
| chr8_+_66070661 | 0.25 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr1_-_171061902 | 0.25 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr19_+_8817883 | 0.24 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr14_-_21102487 | 0.24 |
ENSMUST00000154460.8
ENSMUST00000130291.8 |
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr13_+_58956495 | 0.24 |
ENSMUST00000225950.2
ENSMUST00000225583.2 |
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr6_-_99497900 | 0.24 |
ENSMUST00000176565.8
|
Foxp1
|
forkhead box P1 |
| chr19_-_43974990 | 0.23 |
ENSMUST00000026210.5
|
Cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr2_+_163389068 | 0.23 |
ENSMUST00000109411.8
ENSMUST00000018094.13 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr15_+_31224460 | 0.23 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr9_+_50515207 | 0.23 |
ENSMUST00000044051.6
|
Timm8b
|
translocase of inner mitochondrial membrane 8B |
| chr3_-_108322868 | 0.22 |
ENSMUST00000090558.10
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr11_-_118246332 | 0.22 |
ENSMUST00000017610.10
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr1_+_120048890 | 0.22 |
ENSMUST00000027637.13
ENSMUST00000112644.9 ENSMUST00000056038.15 |
3110009E18Rik
|
RIKEN cDNA 3110009E18 gene |
| chr2_+_158148413 | 0.22 |
ENSMUST00000109491.8
ENSMUST00000016168.9 |
Lbp
|
lipopolysaccharide binding protein |
| chr9_-_58156982 | 0.21 |
ENSMUST00000135310.8
ENSMUST00000085673.11 ENSMUST00000114136.9 ENSMUST00000153820.8 ENSMUST00000124982.2 |
Pml
|
promyelocytic leukemia |
| chr17_+_27236961 | 0.21 |
ENSMUST00000142141.3
ENSMUST00000122106.9 |
Ggnbp1
|
gametogenetin binding protein 1 |
| chr7_-_30259253 | 0.21 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr17_-_89099404 | 0.21 |
ENSMUST00000024916.7
|
Lhcgr
|
luteinizing hormone/choriogonadotropin receptor |
| chr9_+_72892693 | 0.21 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr7_+_142025817 | 0.21 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr19_+_8815024 | 0.20 |
ENSMUST00000159634.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr9_-_58156935 | 0.20 |
ENSMUST00000124063.2
ENSMUST00000126690.8 |
Pml
|
promyelocytic leukemia |
| chr6_-_52181393 | 0.20 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr3_+_89153258 | 0.20 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr9_-_54467419 | 0.20 |
ENSMUST00000041901.7
|
Cib2
|
calcium and integrin binding family member 2 |
| chr5_+_98328723 | 0.20 |
ENSMUST00000112959.4
|
Prdm8
|
PR domain containing 8 |
| chr11_-_69304501 | 0.20 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr7_+_75259778 | 0.20 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr9_+_72946994 | 0.20 |
ENSMUST00000184126.3
|
Pigbos1
|
Pigb opposite strand 1 |
| chr1_-_160134873 | 0.20 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr18_-_78166595 | 0.19 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr9_+_107784065 | 0.19 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr3_-_88366159 | 0.19 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_+_32253016 | 0.19 |
ENSMUST00000132028.8
ENSMUST00000136079.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr5_-_129030367 | 0.19 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr17_-_35392254 | 0.19 |
ENSMUST00000025257.12
|
Aif1
|
allograft inflammatory factor 1 |
| chr5_-_140612993 | 0.19 |
ENSMUST00000199157.2
|
Ttyh3
|
tweety family member 3 |
| chr9_+_72892850 | 0.19 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr4_-_154245073 | 0.19 |
ENSMUST00000105639.4
ENSMUST00000030896.15 |
Tprgl
|
transformation related protein 63 regulated like |
| chr15_+_74828272 | 0.18 |
ENSMUST00000188042.2
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr6_-_97464761 | 0.18 |
ENSMUST00000032146.14
|
Frmd4b
|
FERM domain containing 4B |
| chr13_+_58956077 | 0.18 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr4_+_47091909 | 0.18 |
ENSMUST00000045041.12
ENSMUST00000107744.2 |
Galnt12
|
polypeptide N-acetylgalactosaminyltransferase 12 |
| chr2_+_29759495 | 0.18 |
ENSMUST00000047521.7
ENSMUST00000134152.2 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr7_-_19483389 | 0.18 |
ENSMUST00000108450.5
ENSMUST00000075447.14 |
Nectin2
|
nectin cell adhesion molecule 2 |
| chr3_-_90120942 | 0.18 |
ENSMUST00000195998.5
ENSMUST00000197361.5 ENSMUST00000170122.4 |
Rps27
|
ribosomal protein S27 |
| chr2_+_71811526 | 0.18 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_127050161 | 0.18 |
ENSMUST00000056146.2
|
1810024B03Rik
|
RIKEN cDNA 1810024B03 gene |
| chr9_-_32452885 | 0.18 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr4_-_132149704 | 0.18 |
ENSMUST00000152271.8
ENSMUST00000084170.12 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr11_+_87959067 | 0.17 |
ENSMUST00000018521.11
|
Vezf1
|
vascular endothelial zinc finger 1 |
| chrX_-_77627486 | 0.17 |
ENSMUST00000114025.8
ENSMUST00000134602.8 ENSMUST00000114024.9 |
Prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr3_+_89136353 | 0.17 |
ENSMUST00000041142.4
|
Muc1
|
mucin 1, transmembrane |
| chr15_-_76090508 | 0.17 |
ENSMUST00000073418.13
ENSMUST00000171634.8 ENSMUST00000076442.12 |
Plec
|
plectin |
| chr3_-_137687284 | 0.17 |
ENSMUST00000136613.4
ENSMUST00000029806.13 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr15_-_83054369 | 0.17 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr12_+_84820024 | 0.17 |
ENSMUST00000021667.7
ENSMUST00000222449.2 ENSMUST00000222982.2 |
Isca2
|
iron-sulfur cluster assembly 2 |
| chr13_-_54209669 | 0.17 |
ENSMUST00000021932.6
ENSMUST00000221470.2 |
Drd1
|
dopamine receptor D1 |
| chr7_+_110367375 | 0.17 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr19_+_4282487 | 0.17 |
ENSMUST00000235306.2
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chr19_-_24454720 | 0.17 |
ENSMUST00000099556.2
|
Fam122a
|
family with sequence similarity 122, member A |
| chr13_-_93810808 | 0.17 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr11_+_115044966 | 0.17 |
ENSMUST00000021076.6
|
Rab37
|
RAB37, member RAS oncogene family |
| chr17_+_24946793 | 0.16 |
ENSMUST00000170239.9
|
Rpl3l
|
ribosomal protein L3-like |
| chr7_+_112806672 | 0.16 |
ENSMUST00000047321.9
ENSMUST00000210074.2 ENSMUST00000210238.2 |
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr17_-_57529827 | 0.16 |
ENSMUST00000177425.2
|
C3
|
complement component 3 |
| chr3_-_58792633 | 0.16 |
ENSMUST00000055636.13
ENSMUST00000072551.7 ENSMUST00000051408.8 |
Clrn1
|
clarin 1 |
| chr5_-_135991117 | 0.16 |
ENSMUST00000111150.2
ENSMUST00000054895.4 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
| chrX_+_167819606 | 0.16 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr15_-_81284244 | 0.16 |
ENSMUST00000172107.8
ENSMUST00000169204.2 ENSMUST00000163382.2 |
St13
|
suppression of tumorigenicity 13 |
| chr1_-_120048788 | 0.16 |
ENSMUST00000027634.13
|
Dbi
|
diazepam binding inhibitor |
| chr11_+_33997114 | 0.16 |
ENSMUST00000109329.9
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr15_-_77840856 | 0.16 |
ENSMUST00000117725.2
ENSMUST00000016696.13 |
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr3_-_88277037 | 0.16 |
ENSMUST00000075523.11
|
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr5_-_113957362 | 0.16 |
ENSMUST00000202555.2
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr1_+_74324089 | 0.16 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr4_-_57301791 | 0.16 |
ENSMUST00000075637.11
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr7_+_24990596 | 0.16 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr15_-_76083575 | 0.15 |
ENSMUST00000169438.8
|
Plec
|
plectin |
| chr9_-_106353303 | 0.15 |
ENSMUST00000156426.8
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr19_-_11209797 | 0.15 |
ENSMUST00000186228.3
|
Ms4a12
|
membrane-spanning 4-domains, subfamily A, member 12 |
| chr4_-_155947819 | 0.15 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr16_+_27208660 | 0.15 |
ENSMUST00000143823.2
|
Ccdc50
|
coiled-coil domain containing 50 |
| chr9_+_44245981 | 0.15 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr1_+_166828982 | 0.15 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr3_-_88366351 | 0.15 |
ENSMUST00000165898.8
ENSMUST00000127436.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_+_90511068 | 0.15 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chr8_+_85807566 | 0.15 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr2_-_168432235 | 0.15 |
ENSMUST00000109184.8
|
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr4_+_42950367 | 0.15 |
ENSMUST00000084662.12
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr18_+_61238714 | 0.15 |
ENSMUST00000237706.2
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr12_-_111874489 | 0.15 |
ENSMUST00000054815.15
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr15_-_42540363 | 0.15 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr16_-_92622972 | 0.14 |
ENSMUST00000023673.14
|
Runx1
|
runt related transcription factor 1 |
| chr6_-_124698805 | 0.14 |
ENSMUST00000173315.8
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_-_98329782 | 0.14 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr11_+_29323618 | 0.14 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr1_+_23801007 | 0.14 |
ENSMUST00000063663.6
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr4_-_21767116 | 0.14 |
ENSMUST00000029915.6
|
Tstd3
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 3 |
| chr1_+_59296065 | 0.14 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr7_-_102148780 | 0.14 |
ENSMUST00000216116.4
|
Olfr545
|
olfactory receptor 545 |
| chrX_-_151110425 | 0.14 |
ENSMUST00000195280.3
|
Kantr
|
Kdm5c adjacent non-coding transcript |
| chr4_+_141095415 | 0.14 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr11_+_101046708 | 0.14 |
ENSMUST00000043654.10
|
Tubg2
|
tubulin, gamma 2 |
| chr14_+_55815817 | 0.14 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr4_-_132149780 | 0.14 |
ENSMUST00000102568.10
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr11_-_70350783 | 0.14 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr11_+_78219241 | 0.14 |
ENSMUST00000048073.9
|
Pigs
|
phosphatidylinositol glycan anchor biosynthesis, class S |
| chr2_+_143757193 | 0.14 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr13_-_67053384 | 0.14 |
ENSMUST00000021993.5
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr15_+_54975814 | 0.14 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr10_+_84412490 | 0.14 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr4_+_132968082 | 0.13 |
ENSMUST00000030677.7
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr10_+_93983844 | 0.13 |
ENSMUST00000105290.9
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr12_-_25146078 | 0.13 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chr3_-_88363027 | 0.13 |
ENSMUST00000029700.12
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_-_94492688 | 0.13 |
ENSMUST00000103164.4
|
Acsf2
|
acyl-CoA synthetase family member 2 |
| chr5_-_104125226 | 0.13 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr9_-_106353571 | 0.13 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr3_+_27237143 | 0.13 |
ENSMUST00000091284.5
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr3_+_27237114 | 0.13 |
ENSMUST00000046515.15
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chrX_-_47297746 | 0.13 |
ENSMUST00000088935.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr15_-_77811935 | 0.13 |
ENSMUST00000174529.2
ENSMUST00000173631.8 |
Txn2
|
thioredoxin 2 |
| chrX_-_7765459 | 0.13 |
ENSMUST00000033497.9
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr11_+_4168221 | 0.13 |
ENSMUST00000020699.4
|
Castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr2_+_18069375 | 0.13 |
ENSMUST00000114671.8
ENSMUST00000114680.9 |
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr15_-_74624811 | 0.13 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr15_-_81074921 | 0.13 |
ENSMUST00000131235.9
ENSMUST00000134469.9 ENSMUST00000239114.2 ENSMUST00000149582.8 |
Mrtfa
|
myocardin related transcription factor A |
| chr7_-_29217967 | 0.13 |
ENSMUST00000181975.8
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr4_+_40948407 | 0.13 |
ENSMUST00000030128.6
|
Chmp5
|
charged multivesicular body protein 5 |
| chr7_-_35285001 | 0.13 |
ENSMUST00000069912.6
|
Rgs9bp
|
regulator of G-protein signalling 9 binding protein |
| chr17_-_34340918 | 0.13 |
ENSMUST00000151986.2
|
Brd2
|
bromodomain containing 2 |
| chr17_-_57289121 | 0.13 |
ENSMUST00000056113.5
|
Acer1
|
alkaline ceramidase 1 |
| chr2_+_174126103 | 0.13 |
ENSMUST00000109095.8
ENSMUST00000180362.8 ENSMUST00000109096.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr10_+_127000991 | 0.13 |
ENSMUST00000006914.11
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr9_-_41068771 | 0.13 |
ENSMUST00000136530.8
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chr2_+_38229270 | 0.13 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2 |
| chr7_-_101513300 | 0.12 |
ENSMUST00000106981.8
|
Folr1
|
folate receptor 1 (adult) |
| chr19_+_4281953 | 0.12 |
ENSMUST00000025773.5
|
Pold4
|
polymerase (DNA-directed), delta 4 |
| chrX_-_7765171 | 0.12 |
ENSMUST00000115655.8
ENSMUST00000156741.8 |
Pqbp1
|
polyglutamine binding protein 1 |
| chr8_+_13034245 | 0.12 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr11_-_88608958 | 0.12 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr9_-_85209162 | 0.12 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr13_+_38010879 | 0.12 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr4_-_124587340 | 0.12 |
ENSMUST00000030738.8
|
Utp11
|
UTP11 small subunit processome component |
| chr11_+_105866030 | 0.12 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr10_+_80165961 | 0.12 |
ENSMUST00000186864.7
ENSMUST00000040081.7 |
Reep6
|
receptor accessory protein 6 |
| chr7_+_79939747 | 0.12 |
ENSMUST00000205864.2
|
Vps33b
|
vacuolar protein sorting 33B |
| chr7_+_67925718 | 0.12 |
ENSMUST00000210558.2
|
Fam169b
|
family with sequence similarity 169, member B |
| chr7_-_100613579 | 0.12 |
ENSMUST00000060174.6
|
P2ry6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr12_+_113061819 | 0.12 |
ENSMUST00000109727.9
ENSMUST00000009099.13 ENSMUST00000109723.8 ENSMUST00000109726.8 ENSMUST00000069690.5 |
Mta1
|
metastasis associated 1 |
| chr5_+_124577952 | 0.12 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr7_+_89281897 | 0.12 |
ENSMUST00000032856.13
|
Me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr6_-_33037107 | 0.12 |
ENSMUST00000115091.2
ENSMUST00000127666.8 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.4 | GO:0035854 | eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.3 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.4 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.1 | 0.2 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.2 | GO:0060574 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.5 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:2000872 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.2 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.1 | 0.2 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.2 | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.1 | 0.3 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.0 | 0.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.2 | GO:0042271 | sperm mitochondrion organization(GO:0030382) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0060220 | camera-type eye photoreceptor cell fate commitment(GO:0060220) negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.0 | 0.2 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:2000078 | positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0021679 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0033602 | negative regulation of dopamine secretion(GO:0033602) |
| 0.0 | 0.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 1.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.2 | GO:0048242 | epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.0 | GO:1902566 | regulation of eosinophil degranulation(GO:0043309) regulation of eosinophil activation(GO:1902566) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.0 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) regulation of glucose catabolic process to lactate via pyruvate(GO:1904023) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.0 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.0 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.2 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0046724 | oxalic acid secretion(GO:0046724) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.2 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.2 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 0.3 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.4 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 0.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.2 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 0.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.0 | 0.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-D synthase activity(GO:0004667) prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |