Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
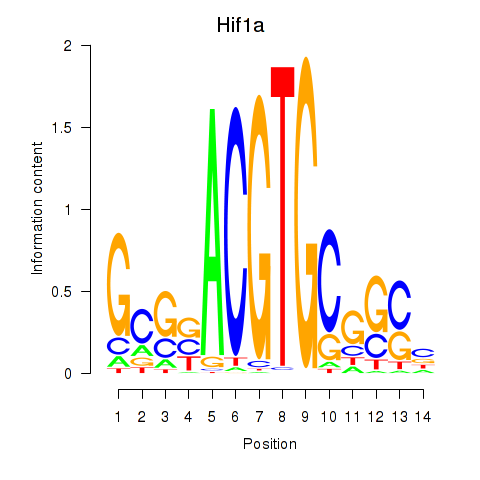
Results for Hif1a
Z-value: 2.12
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSMUSG00000021109.14 | hypoxia inducible factor 1, alpha subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hif1a | mm39_v1_chr12_+_73948143_73948166 | 0.98 | 3.7e-03 | Click! |
Activity profile of Hif1a motif
Sorted Z-values of Hif1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_127178072 | 1.92 |
ENSMUST00000028846.7
|
Dusp2
|
dual specificity phosphatase 2 |
| chr5_+_129097133 | 1.54 |
ENSMUST00000031383.14
ENSMUST00000111343.2 |
Ran
|
RAN, member RAS oncogene family |
| chr7_+_46495256 | 1.31 |
ENSMUST00000048209.16
ENSMUST00000210815.2 ENSMUST00000125862.8 ENSMUST00000210968.2 ENSMUST00000092621.12 ENSMUST00000210467.2 |
Ldha
|
lactate dehydrogenase A |
| chr4_+_150321142 | 1.30 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr7_+_46495521 | 1.12 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A |
| chr2_+_27405169 | 1.04 |
ENSMUST00000113952.10
|
Wdr5
|
WD repeat domain 5 |
| chr14_-_30741012 | 1.03 |
ENSMUST00000037739.8
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr14_-_30740946 | 1.01 |
ENSMUST00000228341.2
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_-_105229653 | 1.00 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr4_+_150321659 | 0.96 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr14_-_31552335 | 0.95 |
ENSMUST00000228037.2
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr1_-_75156993 | 0.92 |
ENSMUST00000027396.15
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr14_+_121148625 | 0.87 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr6_+_85428464 | 0.86 |
ENSMUST00000032078.9
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr11_-_95956176 | 0.85 |
ENSMUST00000100528.5
|
Ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr16_+_32698470 | 0.85 |
ENSMUST00000232272.2
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr2_+_30697838 | 0.84 |
ENSMUST00000041830.10
ENSMUST00000152374.8 |
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr19_+_6952580 | 0.83 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr7_-_98831916 | 0.81 |
ENSMUST00000033001.6
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr10_-_77845571 | 0.81 |
ENSMUST00000020522.9
|
Pfkl
|
phosphofructokinase, liver, B-type |
| chr11_+_120839879 | 0.80 |
ENSMUST00000154187.8
ENSMUST00000100130.10 ENSMUST00000129473.8 ENSMUST00000168579.8 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr5_-_23988551 | 0.79 |
ENSMUST00000148618.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr7_-_126101245 | 0.75 |
ENSMUST00000179818.3
|
Atxn2l
|
ataxin 2-like |
| chr12_+_32870334 | 0.74 |
ENSMUST00000020886.9
|
Nampt
|
nicotinamide phosphoribosyltransferase |
| chr13_-_30168374 | 0.74 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr19_+_6952319 | 0.72 |
ENSMUST00000070850.8
|
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr4_-_129494378 | 0.71 |
ENSMUST00000135055.8
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr5_-_23988696 | 0.69 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr11_+_78069477 | 0.68 |
ENSMUST00000092880.14
ENSMUST00000127587.8 |
Tlcd1
|
TLC domain containing 1 |
| chr15_+_101071948 | 0.63 |
ENSMUST00000000544.12
|
Acvr1b
|
activin A receptor, type 1B |
| chr1_+_172327812 | 0.62 |
ENSMUST00000192460.2
|
Tagln2
|
transgelin 2 |
| chr1_+_16735401 | 0.60 |
ENSMUST00000177501.2
ENSMUST00000065373.6 |
Tmem70
|
transmembrane protein 70 |
| chr8_+_84724130 | 0.59 |
ENSMUST00000095228.5
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr1_-_161078723 | 0.59 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chr16_+_20317544 | 0.59 |
ENSMUST00000003320.14
|
Eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr16_+_20492267 | 0.58 |
ENSMUST00000115460.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr11_+_3282424 | 0.58 |
ENSMUST00000136474.2
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_-_153425791 | 0.57 |
ENSMUST00000041874.9
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr12_+_104998895 | 0.56 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr4_-_129494435 | 0.55 |
ENSMUST00000102593.11
|
Eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr17_-_27352593 | 0.54 |
ENSMUST00000118613.8
|
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr11_+_49916136 | 0.54 |
ENSMUST00000054684.14
ENSMUST00000238748.2 ENSMUST00000102776.5 |
Rnf130
|
ring finger protein 130 |
| chr15_-_75781138 | 0.53 |
ENSMUST00000145764.2
ENSMUST00000116440.9 ENSMUST00000151066.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr1_-_185061525 | 0.53 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr19_+_44994094 | 0.52 |
ENSMUST00000236685.2
|
Twnk
|
twinkle mtDNA helicase |
| chr5_+_45650821 | 0.52 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chrX_-_92875712 | 0.51 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr2_+_162773440 | 0.51 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr6_+_29272625 | 0.51 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr16_+_70110837 | 0.50 |
ENSMUST00000163832.8
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr16_+_70111007 | 0.50 |
ENSMUST00000170464.3
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr7_+_3706992 | 0.50 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chr16_+_18695787 | 0.48 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chr4_-_139079609 | 0.48 |
ENSMUST00000030513.13
ENSMUST00000155257.8 |
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr2_-_14060840 | 0.48 |
ENSMUST00000074854.9
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr6_+_108805594 | 0.47 |
ENSMUST00000089162.5
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr17_-_26314438 | 0.47 |
ENSMUST00000236547.2
|
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr19_+_4560500 | 0.46 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr5_+_124250360 | 0.46 |
ENSMUST00000024470.13
ENSMUST00000119269.6 ENSMUST00000196627.5 ENSMUST00000199457.5 ENSMUST00000198505.2 |
Ogfod2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr17_+_27276262 | 0.46 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr11_-_88608920 | 0.45 |
ENSMUST00000092794.12
|
Msi2
|
musashi RNA-binding protein 2 |
| chr11_+_102080489 | 0.44 |
ENSMUST00000078975.8
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr7_-_138511221 | 0.44 |
ENSMUST00000130500.8
ENSMUST00000106112.2 |
Bnip3
|
BCL2/adenovirus E1B interacting protein 3 |
| chr9_-_96246460 | 0.44 |
ENSMUST00000034983.7
|
Atp1b3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr8_+_107620251 | 0.44 |
ENSMUST00000212272.2
ENSMUST00000047629.7 |
Utp4
|
UTP4 small subunit processome component |
| chr17_-_26314461 | 0.44 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr13_+_32985990 | 0.43 |
ENSMUST00000021832.7
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chrX_+_10583629 | 0.43 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr9_+_107464841 | 0.42 |
ENSMUST00000010192.11
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr5_-_148988413 | 0.42 |
ENSMUST00000093196.11
|
Hmgb1
|
high mobility group box 1 |
| chr1_-_162376053 | 0.41 |
ENSMUST00000028017.16
|
Eef1aknmt
|
EEF1A lysine and N-terminal methyltransferase |
| chr5_+_33815466 | 0.41 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr11_-_87249837 | 0.41 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr1_-_55127183 | 0.40 |
ENSMUST00000027123.15
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr2_+_150590956 | 0.40 |
ENSMUST00000094467.6
|
Entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 |
| chr7_-_44635740 | 0.40 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr2_-_14060774 | 0.40 |
ENSMUST00000114753.8
ENSMUST00000091429.12 |
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr7_+_99808452 | 0.40 |
ENSMUST00000032967.4
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr15_-_75781168 | 0.39 |
ENSMUST00000089680.10
ENSMUST00000141268.8 ENSMUST00000023235.13 ENSMUST00000109972.9 ENSMUST00000089681.12 ENSMUST00000109975.10 ENSMUST00000154584.9 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr5_-_144902598 | 0.39 |
ENSMUST00000110677.8
ENSMUST00000085684.11 ENSMUST00000100461.7 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr17_-_27352876 | 0.39 |
ENSMUST00000119227.3
ENSMUST00000025045.15 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr15_-_75781387 | 0.38 |
ENSMUST00000123712.8
ENSMUST00000141475.2 ENSMUST00000144614.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr6_-_70769135 | 0.38 |
ENSMUST00000066134.6
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr2_+_167922924 | 0.38 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr8_+_34222266 | 0.38 |
ENSMUST00000190675.2
ENSMUST00000171010.8 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr10_+_36850532 | 0.37 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr15_-_57755753 | 0.37 |
ENSMUST00000022993.7
|
Derl1
|
Der1-like domain family, member 1 |
| chr2_-_120970824 | 0.37 |
ENSMUST00000099486.3
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr1_-_55127312 | 0.37 |
ENSMUST00000127861.8
ENSMUST00000144077.3 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr14_+_21102642 | 0.36 |
ENSMUST00000045376.11
|
Adk
|
adenosine kinase |
| chr14_+_21102662 | 0.36 |
ENSMUST00000223915.2
|
Adk
|
adenosine kinase |
| chr9_+_108225026 | 0.36 |
ENSMUST00000035237.12
ENSMUST00000194959.6 |
Usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr8_+_34222058 | 0.35 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr9_-_35028100 | 0.35 |
ENSMUST00000034537.8
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr8_-_46605196 | 0.35 |
ENSMUST00000110378.9
|
Snx25
|
sorting nexin 25 |
| chr2_+_120970979 | 0.34 |
ENSMUST00000119031.8
ENSMUST00000110662.9 |
Adal
|
adenosine deaminase-like |
| chr18_+_35686424 | 0.34 |
ENSMUST00000235679.2
ENSMUST00000235176.2 ENSMUST00000235801.2 ENSMUST00000237592.2 ENSMUST00000237230.2 ENSMUST00000237589.2 |
Snhg4
Snhg4
|
small nucleolar RNA host gene 4 small nucleolar RNA host gene 4 |
| chr7_-_121666486 | 0.34 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr17_+_28988354 | 0.33 |
ENSMUST00000233109.2
ENSMUST00000004986.14 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr10_+_128073900 | 0.33 |
ENSMUST00000105245.3
|
Timeless
|
timeless circadian clock 1 |
| chr19_+_44994905 | 0.33 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr13_-_9815173 | 0.32 |
ENSMUST00000062658.15
ENSMUST00000222358.2 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr4_-_139079842 | 0.32 |
ENSMUST00000102503.10
|
Mrto4
|
mRNA turnover 4, ribosome maturation factor |
| chr9_+_106080307 | 0.32 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr11_+_87017878 | 0.32 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr11_-_79145489 | 0.32 |
ENSMUST00000017821.12
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr6_+_125108829 | 0.32 |
ENSMUST00000044200.11
ENSMUST00000204185.2 |
Nop2
|
NOP2 nucleolar protein |
| chr11_+_60428788 | 0.31 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr13_-_100969823 | 0.30 |
ENSMUST00000225922.2
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr8_+_34221861 | 0.30 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr13_-_9815350 | 0.30 |
ENSMUST00000110636.9
ENSMUST00000152725.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr17_+_73144531 | 0.30 |
ENSMUST00000233886.2
|
Ypel5
|
yippee like 5 |
| chr11_-_115382622 | 0.30 |
ENSMUST00000106530.8
ENSMUST00000021082.7 |
Nt5c
|
5',3'-nucleotidase, cytosolic |
| chr16_+_70110975 | 0.29 |
ENSMUST00000023393.15
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr13_-_9814979 | 0.29 |
ENSMUST00000110634.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr13_+_73752125 | 0.29 |
ENSMUST00000022102.9
|
Clptm1l
|
CLPTM1-like |
| chr11_+_102080446 | 0.28 |
ENSMUST00000070334.10
|
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr7_+_99808526 | 0.28 |
ENSMUST00000207825.2
|
Lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr11_+_72851989 | 0.28 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr8_-_107620210 | 0.27 |
ENSMUST00000177068.8
ENSMUST00000176515.2 ENSMUST00000169312.2 |
Chtf8
Derpc
|
CTF8, chromosome transmission fidelity factor 8 DERPC proline and glycine rich nuclear protein |
| chr16_-_10994135 | 0.27 |
ENSMUST00000037633.16
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr10_+_80062468 | 0.27 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr11_+_87018079 | 0.26 |
ENSMUST00000139532.2
|
Trim37
|
tripartite motif-containing 37 |
| chr4_-_133695264 | 0.25 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr6_+_29272463 | 0.25 |
ENSMUST00000115289.2
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr13_-_9814901 | 0.25 |
ENSMUST00000223421.2
ENSMUST00000128658.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr6_+_88701810 | 0.25 |
ENSMUST00000089449.5
|
Mgll
|
monoglyceride lipase |
| chr17_-_45884179 | 0.24 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr17_-_56933872 | 0.24 |
ENSMUST00000047226.10
|
Lonp1
|
lon peptidase 1, mitochondrial |
| chr11_+_3913970 | 0.24 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr6_-_86503178 | 0.24 |
ENSMUST00000053015.7
|
Pcbp1
|
poly(rC) binding protein 1 |
| chr13_-_9815276 | 0.24 |
ENSMUST00000130151.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr12_-_40087393 | 0.24 |
ENSMUST00000146905.2
|
Arl4a
|
ADP-ribosylation factor-like 4A |
| chr9_+_107446310 | 0.24 |
ENSMUST00000010191.13
ENSMUST00000193747.2 |
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr4_+_150321272 | 0.23 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr13_-_100969878 | 0.23 |
ENSMUST00000067246.6
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr3_-_107993906 | 0.23 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr8_-_122634418 | 0.22 |
ENSMUST00000045557.10
|
Slc7a5
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| chr17_+_46992101 | 0.22 |
ENSMUST00000233612.2
|
Mea1
|
male enhanced antigen 1 |
| chr1_+_74811045 | 0.21 |
ENSMUST00000006716.8
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr12_+_110413523 | 0.21 |
ENSMUST00000222276.2
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_-_44635813 | 0.20 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr11_+_117006020 | 0.20 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr13_-_53135064 | 0.20 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr2_+_120970888 | 0.19 |
ENSMUST00000028702.10
ENSMUST00000110665.8 |
Adal
|
adenosine deaminase-like |
| chr16_-_4238280 | 0.19 |
ENSMUST00000120080.8
|
Adcy9
|
adenylate cyclase 9 |
| chr5_-_123662175 | 0.19 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr18_+_64473091 | 0.19 |
ENSMUST00000175965.10
|
Onecut2
|
one cut domain, family member 2 |
| chr4_-_62438122 | 0.18 |
ENSMUST00000107444.8
ENSMUST00000030090.4 |
Alad
|
aminolevulinate, delta-, dehydratase |
| chr15_-_31453708 | 0.17 |
ENSMUST00000110408.3
|
Ropn1l
|
ropporin 1-like |
| chr17_-_47063095 | 0.17 |
ENSMUST00000121671.2
ENSMUST00000059844.13 |
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr10_+_43354807 | 0.17 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chr2_+_155360015 | 0.16 |
ENSMUST00000103142.12
|
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr19_-_44994824 | 0.16 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr2_-_120971130 | 0.16 |
ENSMUST00000110674.4
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr6_+_88701578 | 0.15 |
ENSMUST00000150180.4
ENSMUST00000163271.8 |
Mgll
|
monoglyceride lipase |
| chr13_-_117162041 | 0.15 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr17_-_23896394 | 0.14 |
ENSMUST00000233428.2
ENSMUST00000233814.2 ENSMUST00000167059.9 ENSMUST00000024698.10 |
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a |
| chr19_-_44058175 | 0.14 |
ENSMUST00000172041.8
ENSMUST00000071698.13 ENSMUST00000112028.10 |
Erlin1
|
ER lipid raft associated 1 |
| chr11_+_117005958 | 0.13 |
ENSMUST00000021177.15
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr1_+_9615619 | 0.13 |
ENSMUST00000072079.9
|
Rrs1
|
ribosome biogenesis regulator 1 |
| chr1_+_63216281 | 0.12 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_+_80591030 | 0.12 |
ENSMUST00000105336.9
|
Dot1l
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae) |
| chr13_-_117161921 | 0.12 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr4_+_116734573 | 0.12 |
ENSMUST00000044823.4
|
Zswim5
|
zinc finger SWIM-type containing 5 |
| chr15_-_102112159 | 0.12 |
ENSMUST00000229252.2
ENSMUST00000229770.2 |
Csad
|
cysteine sulfinic acid decarboxylase |
| chr8_+_46604786 | 0.12 |
ENSMUST00000154040.2
|
Cfap97
|
cilia and flagella associated protein 97 |
| chr5_+_108842294 | 0.12 |
ENSMUST00000013633.12
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr12_+_51394812 | 0.11 |
ENSMUST00000054308.13
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr9_-_88320991 | 0.11 |
ENSMUST00000239462.2
ENSMUST00000165315.9 ENSMUST00000173039.9 |
Snx14
|
sorting nexin 14 |
| chr3_+_94861386 | 0.10 |
ENSMUST00000107260.9
ENSMUST00000142311.8 ENSMUST00000137088.8 ENSMUST00000152869.8 ENSMUST00000107254.8 ENSMUST00000107253.8 |
Rfx5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr13_+_91071077 | 0.10 |
ENSMUST00000051955.9
|
Rps23
|
ribosomal protein S23 |
| chr16_-_18448454 | 0.10 |
ENSMUST00000231622.2
|
Septin5
|
septin 5 |
| chr9_-_88320962 | 0.09 |
ENSMUST00000174806.9
|
Snx14
|
sorting nexin 14 |
| chr6_+_38528738 | 0.09 |
ENSMUST00000161227.8
|
Luc7l2
|
LUC7-like 2 (S. cerevisiae) |
| chr2_+_91357100 | 0.09 |
ENSMUST00000111338.10
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr17_+_46991926 | 0.09 |
ENSMUST00000233491.2
|
Mea1
|
male enhanced antigen 1 |
| chr2_+_164327988 | 0.09 |
ENSMUST00000109350.9
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_121150798 | 0.09 |
ENSMUST00000106113.2
|
Foxk2
|
forkhead box K2 |
| chr2_+_155359868 | 0.08 |
ENSMUST00000029135.15
ENSMUST00000065973.9 |
Acss2
|
acyl-CoA synthetase short-chain family member 2 |
| chr16_-_18448614 | 0.08 |
ENSMUST00000231956.2
ENSMUST00000096987.7 |
Septin5
|
septin 5 |
| chr19_-_44057800 | 0.08 |
ENSMUST00000170801.8
|
Erlin1
|
ER lipid raft associated 1 |
| chr11_-_120239339 | 0.07 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr11_-_116001037 | 0.07 |
ENSMUST00000106441.8
ENSMUST00000021120.6 |
Trim47
|
tripartite motif-containing 47 |
| chr14_-_70680882 | 0.07 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr10_+_43355113 | 0.07 |
ENSMUST00000040147.8
|
Bend3
|
BEN domain containing 3 |
| chr5_-_135280063 | 0.07 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr15_+_81695615 | 0.06 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr6_-_100264439 | 0.06 |
ENSMUST00000101118.4
|
Rybp
|
RING1 and YY1 binding protein |
| chr5_+_14075281 | 0.06 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr4_-_43523595 | 0.05 |
ENSMUST00000107914.10
|
Tpm2
|
tropomyosin 2, beta |
| chr10_+_84938452 | 0.05 |
ENSMUST00000095383.6
|
Tmem263
|
transmembrane protein 263 |
| chr16_+_70110944 | 0.05 |
ENSMUST00000171132.8
|
Gbe1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr14_-_70680659 | 0.05 |
ENSMUST00000180358.3
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr3_-_58599812 | 0.05 |
ENSMUST00000070368.8
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr4_+_129494463 | 0.05 |
ENSMUST00000102591.10
ENSMUST00000181579.8 ENSMUST00000173758.8 |
Tmem234
|
transmembrane protein 234 |
| chr7_+_105290259 | 0.05 |
ENSMUST00000209445.2
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chrX_+_134894573 | 0.05 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr2_-_31973795 | 0.05 |
ENSMUST00000056406.7
|
Fam78a
|
family with sequence similarity 78, member A |
| chr17_+_46991350 | 0.04 |
ENSMUST00000232732.2
|
Mea1
|
male enhanced antigen 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hif1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.3 | 0.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 2.4 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.2 | 0.7 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.2 | 0.7 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.2 | 0.8 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.2 | 0.8 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 0.5 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.2 | 0.5 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.2 | 0.5 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.4 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.4 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.1 | 2.9 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.4 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.1 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.6 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.9 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.5 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.2 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.7 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.9 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.9 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.5 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.3 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 1.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.8 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.4 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) fungiform papilla formation(GO:0061198) |
| 0.1 | 0.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.2 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 1.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.9 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.4 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 1.3 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.0 | 1.3 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.6 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.1 | GO:1904393 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.9 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 1.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 2.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 1.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 2.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 2.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.3 | 0.9 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.2 | 0.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.2 | 0.7 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 0.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.2 | 2.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.8 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 1.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 2.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.5 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.2 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.6 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.8 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.4 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.9 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.8 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.3 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.4 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0001571 | non-tyrosine kinase fibroblast growth factor receptor activity(GO:0001571) |
| 0.0 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.0 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 6.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 3.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |