Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
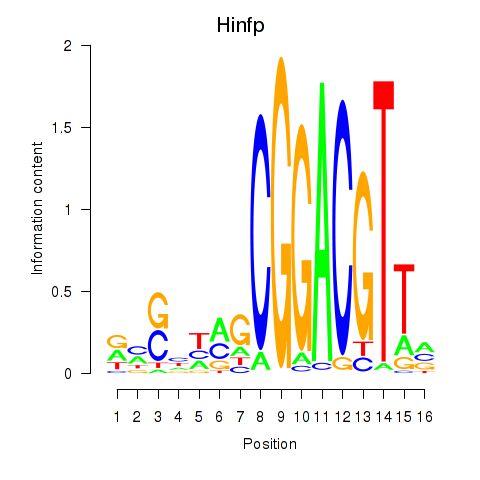
Results for Hinfp
Z-value: 2.04
Transcription factors associated with Hinfp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hinfp
|
ENSMUSG00000032119.6 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hinfp | mm39_v1_chr9_-_44216892_44216986 | 0.42 | 4.8e-01 | Click! |
Activity profile of Hinfp motif
Sorted Z-values of Hinfp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_100165694 | 0.99 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1 |
| chr13_+_9326513 | 0.92 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr5_-_115257336 | 0.92 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr1_-_91340884 | 0.90 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr11_-_88754543 | 0.89 |
ENSMUST00000107904.3
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr5_-_100822097 | 0.88 |
ENSMUST00000031262.9
|
Coq2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr8_-_57940834 | 0.85 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chrX_+_163202778 | 0.84 |
ENSMUST00000208741.2
ENSMUST00000033754.15 ENSMUST00000208697.2 ENSMUST00000208261.2 |
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr5_-_100867520 | 0.83 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr8_+_121395047 | 0.79 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr9_-_55419442 | 0.78 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr5_+_30360246 | 0.76 |
ENSMUST00000026841.15
ENSMUST00000123980.8 ENSMUST00000114783.6 ENSMUST00000114786.8 |
Hadhb
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr17_-_34406193 | 0.73 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr1_+_191449946 | 0.72 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr18_-_33346885 | 0.71 |
ENSMUST00000025236.9
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr5_+_115697526 | 0.71 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chr4_+_150321142 | 0.70 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr13_+_51799268 | 0.70 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr13_-_99027544 | 0.67 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr1_-_16174387 | 0.65 |
ENSMUST00000149566.2
|
Rpl7
|
ribosomal protein L7 |
| chr11_-_101676076 | 0.65 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr3_+_32763313 | 0.63 |
ENSMUST00000126144.3
|
Actl6a
|
actin-like 6A |
| chr3_+_32762656 | 0.63 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A |
| chr7_+_78545756 | 0.63 |
ENSMUST00000107423.2
|
Aen
|
apoptosis enhancing nuclease |
| chr17_+_6130205 | 0.62 |
ENSMUST00000100955.3
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr7_+_78545660 | 0.61 |
ENSMUST00000107425.8
ENSMUST00000107421.8 |
Aen
|
apoptosis enhancing nuclease |
| chr2_+_172841907 | 0.61 |
ENSMUST00000029013.10
ENSMUST00000132212.2 |
Rae1
|
ribonucleic acid export 1 |
| chr6_-_124689094 | 0.61 |
ENSMUST00000004379.8
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr5_-_121590524 | 0.60 |
ENSMUST00000052590.8
ENSMUST00000130451.2 |
Erp29
|
endoplasmic reticulum protein 29 |
| chr5_+_135807334 | 0.60 |
ENSMUST00000019323.11
|
Mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr18_-_36877571 | 0.60 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr11_+_3280771 | 0.58 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr15_+_10981833 | 0.57 |
ENSMUST00000070877.7
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr7_+_46495256 | 0.57 |
ENSMUST00000048209.16
ENSMUST00000210815.2 ENSMUST00000125862.8 ENSMUST00000210968.2 ENSMUST00000092621.12 ENSMUST00000210467.2 |
Ldha
|
lactate dehydrogenase A |
| chr7_-_24997291 | 0.57 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr10_+_81019076 | 0.56 |
ENSMUST00000219133.2
ENSMUST00000047665.7 |
Dapk3
|
death-associated protein kinase 3 |
| chr6_-_113717689 | 0.55 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr12_+_100076407 | 0.54 |
ENSMUST00000021595.10
|
Psmc1
|
protease (prosome, macropain) 26S subunit, ATPase 1 |
| chr4_+_152123772 | 0.53 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr7_+_46495521 | 0.53 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A |
| chr2_-_156848923 | 0.52 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr11_-_31621863 | 0.52 |
ENSMUST00000058060.14
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr7_-_121666486 | 0.52 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr9_+_54858388 | 0.52 |
ENSMUST00000171900.2
|
Psma4
|
proteasome subunit alpha 4 |
| chr11_+_58221538 | 0.52 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr14_-_30723549 | 0.52 |
ENSMUST00000226782.2
ENSMUST00000186131.7 ENSMUST00000228767.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr17_-_80514725 | 0.51 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr5_-_137785903 | 0.51 |
ENSMUST00000196022.2
|
Mepce
|
methylphosphate capping enzyme |
| chr15_-_31601652 | 0.50 |
ENSMUST00000161266.2
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr10_+_81019117 | 0.50 |
ENSMUST00000218157.2
|
Dapk3
|
death-associated protein kinase 3 |
| chr11_-_62172164 | 0.49 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr3_-_88857578 | 0.49 |
ENSMUST00000174402.8
ENSMUST00000174077.8 |
Dap3
|
death associated protein 3 |
| chr19_-_6168518 | 0.48 |
ENSMUST00000113533.3
|
Sac3d1
|
SAC3 domain containing 1 |
| chr6_-_39396691 | 0.48 |
ENSMUST00000146785.8
ENSMUST00000114823.8 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr12_+_70499869 | 0.47 |
ENSMUST00000021471.13
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr4_-_40757814 | 0.47 |
ENSMUST00000030117.5
|
Smu1
|
smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) |
| chr8_+_124204598 | 0.47 |
ENSMUST00000001520.13
|
Afg3l1
|
AFG3-like AAA ATPase 1 |
| chr9_+_73020708 | 0.47 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr19_+_45549009 | 0.47 |
ENSMUST00000047057.9
|
Gm17018
|
predicted gene 17018 |
| chr6_+_29272625 | 0.46 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chrX_+_163202852 | 0.46 |
ENSMUST00000112255.8
|
Piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr8_+_121394961 | 0.45 |
ENSMUST00000034276.13
ENSMUST00000181586.8 |
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr3_+_87826834 | 0.45 |
ENSMUST00000137775.2
|
Mrpl24
|
mitochondrial ribosomal protein L24 |
| chr1_-_171050004 | 0.45 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr13_+_41154478 | 0.45 |
ENSMUST00000046951.10
|
Pak1ip1
|
PAK1 interacting protein 1 |
| chrX_-_74460168 | 0.44 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr8_+_85786684 | 0.44 |
ENSMUST00000095220.4
|
Fbxw9
|
F-box and WD-40 domain protein 9 |
| chr18_-_35087355 | 0.44 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr5_+_135807528 | 0.44 |
ENSMUST00000200556.5
ENSMUST00000196285.2 |
Mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr6_-_124689001 | 0.44 |
ENSMUST00000203238.2
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr5_+_115417725 | 0.43 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr16_-_31821938 | 0.43 |
ENSMUST00000023457.13
ENSMUST00000231360.2 |
Senp5
|
SUMO/sentrin specific peptidase 5 |
| chr11_+_58221569 | 0.43 |
ENSMUST00000073128.7
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr4_+_128887017 | 0.43 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr9_+_20914211 | 0.43 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr6_+_47930324 | 0.43 |
ENSMUST00000101445.11
|
Zfp956
|
zinc finger protein 956 |
| chr13_+_21663077 | 0.43 |
ENSMUST00000062609.6
ENSMUST00000225845.2 |
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr11_+_100973391 | 0.42 |
ENSMUST00000001806.10
ENSMUST00000107308.4 |
Coasy
|
Coenzyme A synthase |
| chr1_+_179495767 | 0.42 |
ENSMUST00000040538.10
|
Sccpdh
|
saccharopine dehydrogenase (putative) |
| chr16_-_94327689 | 0.42 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr16_-_4782031 | 0.42 |
ENSMUST00000023157.6
ENSMUST00000229765.2 |
Anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr4_+_156194427 | 0.41 |
ENSMUST00000072554.13
ENSMUST00000169550.8 ENSMUST00000105576.2 |
9430015G10Rik
|
RIKEN cDNA 9430015G10 gene |
| chr14_-_30723292 | 0.41 |
ENSMUST00000228736.2
ENSMUST00000226374.2 |
Spcs1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr8_+_75742850 | 0.41 |
ENSMUST00000109940.2
|
Hmgxb4
|
HMG box domain containing 4 |
| chr3_+_90138895 | 0.40 |
ENSMUST00000029546.15
ENSMUST00000119304.2 |
Jtb
|
jumping translocation breakpoint |
| chr11_-_79145489 | 0.40 |
ENSMUST00000017821.12
|
Wsb1
|
WD repeat and SOCS box-containing 1 |
| chr4_-_119279551 | 0.40 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr2_+_150751475 | 0.40 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr11_-_117859997 | 0.40 |
ENSMUST00000054002.4
|
Socs3
|
suppressor of cytokine signaling 3 |
| chr1_+_191553556 | 0.40 |
ENSMUST00000027931.8
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr15_-_79430742 | 0.40 |
ENSMUST00000231053.2
ENSMUST00000229431.2 |
Ddx17
|
DEAD box helicase 17 |
| chr9_+_59564482 | 0.39 |
ENSMUST00000216620.2
ENSMUST00000217038.2 |
Pkm
|
pyruvate kinase, muscle |
| chr8_+_84442133 | 0.38 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a |
| chr8_+_27532623 | 0.38 |
ENSMUST00000209856.2
ENSMUST00000098851.12 ENSMUST00000211393.2 ENSMUST00000211518.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr17_+_64244946 | 0.38 |
ENSMUST00000038080.7
|
Fer
|
fer (fms/fps related) protein kinase |
| chr14_-_25927674 | 0.38 |
ENSMUST00000100811.6
|
Tmem254a
|
transmembrane protein 254a |
| chr7_-_27713540 | 0.38 |
ENSMUST00000180502.8
|
Zfp850
|
zinc finger protein 850 |
| chr3_-_65300000 | 0.37 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr2_-_127286444 | 0.37 |
ENSMUST00000028848.4
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr14_+_20344765 | 0.36 |
ENSMUST00000223663.2
ENSMUST00000022343.6 ENSMUST00000224066.2 ENSMUST00000223941.2 ENSMUST00000224311.2 |
Nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr6_-_47790272 | 0.36 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr2_+_5850053 | 0.36 |
ENSMUST00000127116.7
ENSMUST00000194933.2 |
Nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr4_+_124608569 | 0.36 |
ENSMUST00000030734.5
|
Sf3a3
|
splicing factor 3a, subunit 3 |
| chrY_+_1010543 | 0.36 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr7_+_97049210 | 0.36 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr8_-_112026854 | 0.35 |
ENSMUST00000038739.5
|
Rfwd3
|
ring finger and WD repeat domain 3 |
| chr4_-_133695264 | 0.35 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr9_+_57468217 | 0.35 |
ENSMUST00000045791.11
ENSMUST00000216986.2 |
Scamp2
|
secretory carrier membrane protein 2 |
| chr19_-_45548942 | 0.35 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chr19_+_4560500 | 0.34 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr16_+_4501934 | 0.34 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr1_+_185095232 | 0.34 |
ENSMUST00000046514.13
|
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr13_-_99027482 | 0.34 |
ENSMUST00000179301.8
ENSMUST00000179271.2 |
Tnpo1
|
transportin 1 |
| chr11_-_115405200 | 0.34 |
ENSMUST00000021083.7
|
Jpt1
|
Jupiter microtubule associated homolog 1 |
| chr3_-_88857213 | 0.33 |
ENSMUST00000172942.8
ENSMUST00000107491.11 |
Dap3
|
death associated protein 3 |
| chr11_+_101556367 | 0.33 |
ENSMUST00000039388.3
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chr1_-_171050077 | 0.33 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr16_+_35861554 | 0.33 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr3_-_138780894 | 0.33 |
ENSMUST00000196280.5
ENSMUST00000200396.2 |
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chr6_-_50543514 | 0.32 |
ENSMUST00000161401.2
|
Cycs
|
cytochrome c, somatic |
| chr11_+_76836330 | 0.32 |
ENSMUST00000021197.10
|
Blmh
|
bleomycin hydrolase |
| chr1_+_131455635 | 0.32 |
ENSMUST00000068613.5
|
Fam72a
|
family with sequence similarity 72, member A |
| chr8_-_61436249 | 0.31 |
ENSMUST00000004430.14
ENSMUST00000110301.2 ENSMUST00000093490.9 |
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr2_+_5849828 | 0.31 |
ENSMUST00000026927.10
ENSMUST00000179748.8 |
Nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr18_+_35695339 | 0.30 |
ENSMUST00000237365.2
|
Matr3
|
matrin 3 |
| chr8_-_110419867 | 0.30 |
ENSMUST00000034164.6
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr9_+_108167628 | 0.30 |
ENSMUST00000035227.8
|
Nicn1
|
nicolin 1 |
| chr4_-_116982804 | 0.29 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr15_-_35938155 | 0.29 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr3_-_88857707 | 0.29 |
ENSMUST00000090938.11
|
Dap3
|
death associated protein 3 |
| chr17_+_6130061 | 0.29 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr6_-_119825081 | 0.29 |
ENSMUST00000183703.8
ENSMUST00000183911.8 |
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr2_+_29014984 | 0.29 |
ENSMUST00000061578.9
|
Setx
|
senataxin |
| chr11_+_17161912 | 0.29 |
ENSMUST00000046955.7
|
Wdr92
|
WD repeat domain 92 |
| chr1_-_171854818 | 0.28 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr19_-_27407206 | 0.28 |
ENSMUST00000076219.6
|
Pum3
|
pumilio RNA-binding family member 3 |
| chr9_-_66956425 | 0.28 |
ENSMUST00000113687.8
ENSMUST00000113693.8 ENSMUST00000113701.8 ENSMUST00000034928.12 ENSMUST00000113685.10 ENSMUST00000030185.5 ENSMUST00000050905.16 ENSMUST00000113705.8 ENSMUST00000113697.8 ENSMUST00000113707.9 |
Tpm1
|
tropomyosin 1, alpha |
| chr11_-_106890307 | 0.28 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr11_-_31621727 | 0.28 |
ENSMUST00000109415.2
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr6_+_121160626 | 0.27 |
ENSMUST00000118234.8
ENSMUST00000088561.10 ENSMUST00000137432.8 ENSMUST00000120066.8 |
Pex26
|
peroxisomal biogenesis factor 26 |
| chr5_+_137777111 | 0.27 |
ENSMUST00000126126.8
ENSMUST00000031739.6 |
Ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr11_-_120524362 | 0.27 |
ENSMUST00000058162.14
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr8_-_25506756 | 0.27 |
ENSMUST00000084032.6
ENSMUST00000207132.2 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr7_-_44578834 | 0.27 |
ENSMUST00000107857.11
ENSMUST00000167930.8 ENSMUST00000085399.13 ENSMUST00000166972.9 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr2_+_75489596 | 0.26 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr5_+_33815910 | 0.26 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr15_-_79430942 | 0.26 |
ENSMUST00000054014.9
ENSMUST00000229877.2 |
Ddx17
|
DEAD box helicase 17 |
| chrX_+_73314418 | 0.26 |
ENSMUST00000008826.14
ENSMUST00000151702.8 ENSMUST00000074085.12 ENSMUST00000135690.2 |
Rpl10
|
ribosomal protein L10 |
| chr3_+_60910207 | 0.26 |
ENSMUST00000029331.7
ENSMUST00000193201.2 ENSMUST00000193943.2 |
P2ry1
|
purinergic receptor P2Y, G-protein coupled 1 |
| chr19_-_45738002 | 0.26 |
ENSMUST00000070215.8
|
Npm3
|
nucleoplasmin 3 |
| chr12_-_110662765 | 0.26 |
ENSMUST00000094361.11
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr5_-_8472696 | 0.26 |
ENSMUST00000171808.8
|
Dbf4
|
DBF4 zinc finger |
| chr2_+_92205651 | 0.26 |
ENSMUST00000028650.9
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr2_+_28423367 | 0.25 |
ENSMUST00000113893.8
ENSMUST00000100241.10 |
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr5_-_8472582 | 0.25 |
ENSMUST00000168500.8
ENSMUST00000002368.16 |
Dbf4
|
DBF4 zinc finger |
| chr17_-_23892798 | 0.25 |
ENSMUST00000047436.11
ENSMUST00000115490.9 ENSMUST00000095579.11 |
Thoc6
|
THO complex 6 |
| chr19_+_41900360 | 0.25 |
ENSMUST00000011896.8
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr6_+_29272463 | 0.25 |
ENSMUST00000115289.2
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chrX_-_73397181 | 0.25 |
ENSMUST00000114152.2
ENSMUST00000114153.2 ENSMUST00000015433.4 |
Lage3
|
L antigen family, member 3 |
| chr5_+_33815892 | 0.25 |
ENSMUST00000152847.8
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_+_119378178 | 0.25 |
ENSMUST00000014221.13
ENSMUST00000119172.2 |
Chp1
|
calcineurin-like EF hand protein 1 |
| chr3_+_103078971 | 0.25 |
ENSMUST00000005830.15
|
Bcas2
|
breast carcinoma amplified sequence 2 |
| chr7_+_66339637 | 0.25 |
ENSMUST00000153007.2
ENSMUST00000121777.9 ENSMUST00000150071.8 ENSMUST00000077967.13 |
Lins1
|
lines homolog 1 |
| chr3_-_105594865 | 0.25 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr5_-_30360113 | 0.25 |
ENSMUST00000156859.3
|
Hadha
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha |
| chr9_-_36678868 | 0.24 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr4_-_62443273 | 0.24 |
ENSMUST00000030091.10
|
Pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr6_-_106777014 | 0.24 |
ENSMUST00000013882.10
ENSMUST00000113239.10 |
Crbn
|
cereblon |
| chr19_-_43512929 | 0.24 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr18_-_56695333 | 0.24 |
ENSMUST00000066208.13
ENSMUST00000172734.8 |
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr1_+_91468796 | 0.24 |
ENSMUST00000188081.7
ENSMUST00000188879.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chrX_-_74460137 | 0.23 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr9_-_105372235 | 0.23 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr9_+_54858092 | 0.23 |
ENSMUST00000172407.8
|
Psma4
|
proteasome subunit alpha 4 |
| chr7_-_19595221 | 0.23 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr11_+_120604804 | 0.23 |
ENSMUST00000151852.2
|
Lrrc45
|
leucine rich repeat containing 45 |
| chr7_+_112553162 | 0.23 |
ENSMUST00000182858.2
|
Rassf10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10 |
| chr11_+_83189831 | 0.22 |
ENSMUST00000176944.8
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr12_+_116369017 | 0.22 |
ENSMUST00000084828.5
ENSMUST00000222469.2 ENSMUST00000221114.2 ENSMUST00000221970.2 |
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr9_-_59393893 | 0.22 |
ENSMUST00000171975.8
|
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr8_+_84441806 | 0.22 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a |
| chr12_+_111132779 | 0.22 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr14_+_30547541 | 0.22 |
ENSMUST00000006701.8
|
Stimate
|
STIM activating enhancer |
| chr16_-_4536992 | 0.22 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1 |
| chr4_+_155915729 | 0.21 |
ENSMUST00000139651.8
ENSMUST00000084097.12 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr9_+_108660989 | 0.20 |
ENSMUST00000192307.6
ENSMUST00000193560.6 ENSMUST00000194875.6 |
Ip6k2
|
inositol hexaphosphate kinase 2 |
| chr17_+_28491085 | 0.20 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr8_+_84441854 | 0.20 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a |
| chr10_+_13428638 | 0.19 |
ENSMUST00000019944.9
|
Adat2
|
adenosine deaminase, tRNA-specific 2 |
| chr1_-_97589675 | 0.19 |
ENSMUST00000053033.14
ENSMUST00000149927.2 |
Macir
|
macrophage immunometabolism regulator |
| chr11_-_48707763 | 0.19 |
ENSMUST00000140800.2
|
Trim41
|
tripartite motif-containing 41 |
| chr9_-_105372677 | 0.19 |
ENSMUST00000176036.8
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr7_-_92523396 | 0.19 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr11_-_73029070 | 0.19 |
ENSMUST00000052140.3
|
Haspin
|
histone H3 associated protein kinase |
| chr7_+_121758646 | 0.19 |
ENSMUST00000033154.8
ENSMUST00000205901.2 |
Plk1
|
polo like kinase 1 |
| chr15_-_31453708 | 0.18 |
ENSMUST00000110408.3
|
Ropn1l
|
ropporin 1-like |
| chr4_-_106321363 | 0.18 |
ENSMUST00000049507.6
|
Pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr5_-_137305895 | 0.18 |
ENSMUST00000199243.5
ENSMUST00000197466.5 ENSMUST00000040873.12 |
Srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr17_-_36227199 | 0.18 |
ENSMUST00000172642.2
ENSMUST00000174807.8 ENSMUST00000174349.8 ENSMUST00000025305.16 ENSMUST00000113782.10 |
Mrps18b
|
mitochondrial ribosomal protein S18B |
| chr17_+_27136065 | 0.18 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hinfp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.3 | 0.8 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.3 | 1.0 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.2 | 0.8 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 0.2 | GO:0032805 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 0.2 | 0.7 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.2 | 0.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 0.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.2 | GO:0043519 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 | 0.7 | GO:1904253 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.5 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.1 | 0.4 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 1.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.5 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.1 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.9 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.3 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.3 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 1.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.4 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.6 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.1 | 0.3 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 0.5 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.3 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.5 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.2 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 0.1 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 1.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.3 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.5 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 0.4 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.4 | GO:0038095 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.6 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.3 | GO:1902031 | regulation of pentose-phosphate shunt(GO:0043456) regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.0 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.0 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.4 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.0 | 0.7 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.2 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.0 | 0.5 | GO:1904871 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0042994 | astral microtubule organization(GO:0030953) cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.7 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.1 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.1 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 1.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.5 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.8 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.4 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.9 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.2 | GO:1990667 | PCSK9-LDLR complex(GO:1990666) PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 2.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:1990391 | DNA repair complex(GO:1990391) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 1.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.2 | 0.7 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.2 | 1.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.5 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 0.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.3 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0072354 | histone kinase activity (H3-T3 specific)(GO:0072354) |
| 0.0 | 0.3 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.6 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.7 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.0 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |