Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
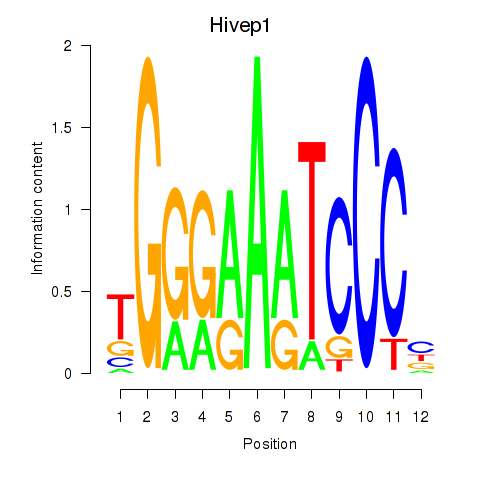
Results for Hivep1
Z-value: 1.25
Transcription factors associated with Hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hivep1
|
ENSMUSG00000021366.9 | human immunodeficiency virus type I enhancer binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hivep1 | mm39_v1_chr13_+_42205790_42205880 | -0.24 | 7.0e-01 | Click! |
Activity profile of Hivep1 motif
Sorted Z-values of Hivep1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_129252396 | 2.22 |
ENSMUST00000032259.6
|
Cd69
|
CD69 antigen |
| chr17_+_35598583 | 1.41 |
ENSMUST00000081435.5
|
H2-Q4
|
histocompatibility 2, Q region locus 4 |
| chr6_-_129252323 | 1.19 |
ENSMUST00000204411.2
|
Cd69
|
CD69 antigen |
| chr17_-_34219225 | 1.01 |
ENSMUST00000238098.2
ENSMUST00000087189.7 ENSMUST00000173075.3 ENSMUST00000172912.8 ENSMUST00000236740.2 ENSMUST00000025181.18 |
H2-K1
|
histocompatibility 2, K1, K region |
| chr14_-_55828511 | 0.91 |
ENSMUST00000161807.8
ENSMUST00000111378.10 ENSMUST00000159687.2 |
Psme2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr12_-_55539372 | 0.82 |
ENSMUST00000021413.9
|
Nfkbia
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha |
| chr5_-_138169253 | 0.79 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr5_-_138169509 | 0.72 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr17_+_35780977 | 0.66 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr6_-_83030759 | 0.60 |
ENSMUST00000134606.8
|
Htra2
|
HtrA serine peptidase 2 |
| chr5_-_92496730 | 0.58 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr11_+_101207743 | 0.58 |
ENSMUST00000151385.2
|
Psme3
|
proteaseome (prosome, macropain) activator subunit 3 (PA28 gamma, Ki) |
| chr17_+_35482063 | 0.54 |
ENSMUST00000172503.3
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr18_+_84106796 | 0.51 |
ENSMUST00000235383.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr18_+_84106188 | 0.50 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr7_-_46365108 | 0.50 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr17_-_28736483 | 0.50 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr4_-_40239778 | 0.50 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr7_-_30855295 | 0.44 |
ENSMUST00000186723.3
|
Gramd1a
|
GRAM domain containing 1A |
| chr17_+_35481702 | 0.43 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr4_-_43031370 | 0.43 |
ENSMUST00000138030.2
ENSMUST00000136326.8 |
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr17_-_34109513 | 0.43 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr17_+_75024727 | 0.43 |
ENSMUST00000024882.8
ENSMUST00000234751.2 ENSMUST00000234568.2 |
Ttc27
|
tetratricopeptide repeat domain 27 |
| chr4_+_123176570 | 0.42 |
ENSMUST00000106243.8
ENSMUST00000106241.8 ENSMUST00000080178.13 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr3_+_127584251 | 0.40 |
ENSMUST00000164447.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr12_-_40298072 | 0.39 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr3_-_88858465 | 0.39 |
ENSMUST00000173135.8
|
Dap3
|
death associated protein 3 |
| chr3_+_115681788 | 0.38 |
ENSMUST00000196804.5
ENSMUST00000106505.8 ENSMUST00000043342.10 |
Dph5
|
diphthamide biosynthesis 5 |
| chr15_+_76594810 | 0.37 |
ENSMUST00000136840.8
ENSMUST00000127208.8 ENSMUST00000036423.15 ENSMUST00000137649.8 ENSMUST00000155225.2 ENSMUST00000155735.2 |
Lrrc14
|
leucine rich repeat containing 14 |
| chr11_+_70548022 | 0.35 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr14_+_55798362 | 0.34 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_-_138169476 | 0.34 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr11_+_97697328 | 0.34 |
ENSMUST00000153520.3
|
Lasp1
|
LIM and SH3 protein 1 |
| chr11_-_5492175 | 0.33 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr17_-_35081456 | 0.33 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr15_+_102379503 | 0.32 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr3_-_88858402 | 0.31 |
ENSMUST00000173021.8
|
Dap3
|
death associated protein 3 |
| chr14_+_52222283 | 0.31 |
ENSMUST00000093813.12
ENSMUST00000100639.11 ENSMUST00000182909.8 ENSMUST00000182760.8 ENSMUST00000182061.8 ENSMUST00000182193.2 |
Arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr19_+_24853039 | 0.29 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr2_-_93292257 | 0.29 |
ENSMUST00000150508.8
|
Cd82
|
CD82 antigen |
| chr4_-_40239700 | 0.28 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr1_+_161796854 | 0.28 |
ENSMUST00000160881.2
ENSMUST00000159648.2 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr11_+_60590584 | 0.28 |
ENSMUST00000108719.4
|
Llgl1
|
LLGL1 scribble cell polarity complex component |
| chr12_-_113912416 | 0.27 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chr14_+_55798517 | 0.27 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr7_+_27305222 | 0.27 |
ENSMUST00000143499.8
ENSMUST00000108343.8 |
Akt2
|
thymoma viral proto-oncogene 2 |
| chr9_+_95739650 | 0.26 |
ENSMUST00000034980.9
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr9_+_5308828 | 0.26 |
ENSMUST00000162846.8
ENSMUST00000027012.14 |
Casp4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr3_-_107667499 | 0.26 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr9_+_95739747 | 0.26 |
ENSMUST00000215311.2
|
Atr
|
ataxia telangiectasia and Rad3 related |
| chr3_-_65865656 | 0.24 |
ENSMUST00000029416.14
|
Ccnl1
|
cyclin L1 |
| chr7_+_27305271 | 0.23 |
ENSMUST00000051356.12
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr2_-_26917921 | 0.23 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr18_-_34784746 | 0.23 |
ENSMUST00000025228.12
ENSMUST00000133181.2 |
Cdc23
|
CDC23 cell division cycle 23 |
| chr17_+_27136065 | 0.22 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr4_+_151012375 | 0.22 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr7_-_125799585 | 0.22 |
ENSMUST00000163959.2
|
Xpo6
|
exportin 6 |
| chr11_-_97913420 | 0.22 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr7_-_19363280 | 0.21 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr5_-_36771074 | 0.21 |
ENSMUST00000132383.6
ENSMUST00000174019.2 |
D5Ertd579e
Gm42936
|
DNA segment, Chr 5, ERATO Doi 579, expressed predicted gene 42936 |
| chr4_+_126156118 | 0.20 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr12_+_111132779 | 0.20 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr2_-_126341757 | 0.20 |
ENSMUST00000040128.12
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr16_-_18052937 | 0.19 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr2_-_5068807 | 0.19 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr13_-_100382831 | 0.19 |
ENSMUST00000049789.3
|
Naip5
|
NLR family, apoptosis inhibitory protein 5 |
| chr7_+_79836581 | 0.18 |
ENSMUST00000032754.9
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr17_+_35413415 | 0.18 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr2_+_163535925 | 0.18 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr17_+_71511642 | 0.17 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr16_-_3725515 | 0.17 |
ENSMUST00000177221.2
ENSMUST00000177323.8 |
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chr7_+_99184645 | 0.17 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr16_-_52272828 | 0.16 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr9_-_57672124 | 0.16 |
ENSMUST00000215158.2
|
Clk3
|
CDC-like kinase 3 |
| chr12_+_111132847 | 0.16 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3 |
| chr1_-_36312482 | 0.16 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr1_+_55276336 | 0.16 |
ENSMUST00000061334.10
|
Mars2
|
methionine-tRNA synthetase 2 (mitochondrial) |
| chr9_-_44646487 | 0.16 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_-_70128712 | 0.16 |
ENSMUST00000108577.8
ENSMUST00000108579.8 ENSMUST00000021181.13 ENSMUST00000108578.9 ENSMUST00000102569.10 |
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr9_-_78389006 | 0.14 |
ENSMUST00000042235.15
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr2_+_118730823 | 0.14 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr11_+_94102255 | 0.14 |
ENSMUST00000041589.6
|
Tob1
|
transducer of ErbB-2.1 |
| chr15_+_102379621 | 0.14 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr14_+_55797934 | 0.14 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr17_-_84495364 | 0.13 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr2_-_5068743 | 0.13 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr6_+_83034825 | 0.12 |
ENSMUST00000077502.5
|
Dqx1
|
DEAQ RNA-dependent ATPase |
| chr11_-_101193032 | 0.12 |
ENSMUST00000140706.8
ENSMUST00000170502.2 ENSMUST00000172233.8 |
Becn1
|
beclin 1, autophagy related |
| chr11_+_78237492 | 0.11 |
ENSMUST00000100755.4
|
Unc119
|
unc-119 lipid binding chaperone |
| chrX_-_133501677 | 0.10 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr8_+_61085890 | 0.10 |
ENSMUST00000160719.8
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr5_-_144902598 | 0.10 |
ENSMUST00000110677.8
ENSMUST00000085684.11 ENSMUST00000100461.7 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr11_+_100751151 | 0.10 |
ENSMUST00000138083.8
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr3_+_127584449 | 0.10 |
ENSMUST00000171621.3
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr13_+_43938251 | 0.10 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr19_-_6919755 | 0.09 |
ENSMUST00000099782.10
|
Gpr137
|
G protein-coupled receptor 137 |
| chr5_-_98178834 | 0.09 |
ENSMUST00000199088.2
|
Antxr2
|
anthrax toxin receptor 2 |
| chr11_-_93846453 | 0.09 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr7_+_30121776 | 0.09 |
ENSMUST00000108175.8
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr4_-_133695264 | 0.09 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_172418058 | 0.09 |
ENSMUST00000065679.8
|
Slamf8
|
SLAM family member 8 |
| chr4_+_155646807 | 0.09 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chrX_-_133501874 | 0.08 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr6_+_55313409 | 0.08 |
ENSMUST00000004774.4
|
Aqp1
|
aquaporin 1 |
| chr4_-_82423985 | 0.08 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr17_+_45088030 | 0.08 |
ENSMUST00000050630.14
|
Supt3
|
SPT3, SAGA and STAGA complex component |
| chr1_+_192984278 | 0.08 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr11_+_78079562 | 0.08 |
ENSMUST00000108322.9
|
Rab34
|
RAB34, member RAS oncogene family |
| chr6_-_8778439 | 0.08 |
ENSMUST00000115520.8
ENSMUST00000038403.12 ENSMUST00000115518.8 |
Ica1
|
islet cell autoantigen 1 |
| chr12_+_85733415 | 0.08 |
ENSMUST00000040536.6
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr4_-_120672900 | 0.08 |
ENSMUST00000120779.8
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr1_+_161796748 | 0.08 |
ENSMUST00000111594.3
ENSMUST00000028021.7 ENSMUST00000193784.2 ENSMUST00000161826.2 |
Pigc
Gm38304
|
phosphatidylinositol glycan anchor biosynthesis, class C predicted gene, 38304 |
| chr11_+_78079631 | 0.07 |
ENSMUST00000056241.12
ENSMUST00000207728.2 |
Rab34
|
RAB34, member RAS oncogene family |
| chr17_-_36012932 | 0.07 |
ENSMUST00000166980.9
ENSMUST00000145900.8 |
Ddr1
|
discoidin domain receptor family, member 1 |
| chr5_-_98178811 | 0.07 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr17_+_45866618 | 0.07 |
ENSMUST00000024742.9
ENSMUST00000233929.2 |
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon |
| chr5_+_30437579 | 0.07 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chr13_-_19521337 | 0.06 |
ENSMUST00000103563.3
|
Trgv2
|
T cell receptor gamma variable 2 |
| chr7_-_125799644 | 0.06 |
ENSMUST00000168189.8
|
Xpo6
|
exportin 6 |
| chr12_-_113823290 | 0.06 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr15_+_78129040 | 0.06 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr14_+_51045298 | 0.06 |
ENSMUST00000036126.7
|
Parp2
|
poly (ADP-ribose) polymerase family, member 2 |
| chr3_+_96508400 | 0.06 |
ENSMUST00000062058.5
|
Lix1l
|
Lix1-like |
| chr17_+_56312672 | 0.06 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr6_-_48818006 | 0.05 |
ENSMUST00000203229.3
|
Tmem176b
|
transmembrane protein 176B |
| chr8_-_47866869 | 0.05 |
ENSMUST00000211882.2
|
Stox2
|
storkhead box 2 |
| chr1_+_186947934 | 0.05 |
ENSMUST00000160471.8
|
Gpatch2
|
G patch domain containing 2 |
| chr12_-_113945961 | 0.05 |
ENSMUST00000103466.2
|
Ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr7_+_30121147 | 0.05 |
ENSMUST00000108176.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr6_-_8778106 | 0.05 |
ENSMUST00000151758.2
ENSMUST00000115519.8 ENSMUST00000153390.8 |
Ica1
|
islet cell autoantigen 1 |
| chr12_-_28685849 | 0.04 |
ENSMUST00000221871.2
|
Rps7
|
ribosomal protein S7 |
| chr11_+_75546989 | 0.04 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr5_-_92231517 | 0.04 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_+_127864847 | 0.04 |
ENSMUST00000118169.8
ENSMUST00000206909.2 ENSMUST00000142841.8 |
Slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr7_-_125799570 | 0.04 |
ENSMUST00000009344.16
|
Xpo6
|
exportin 6 |
| chr11_-_70128678 | 0.04 |
ENSMUST00000108575.9
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr11_+_101066867 | 0.04 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
| chr6_+_41090484 | 0.03 |
ENSMUST00000103267.3
|
Trbv12-1
|
T cell receptor beta, variable 12-1 |
| chr12_+_98886826 | 0.03 |
ENSMUST00000085109.10
ENSMUST00000079146.13 |
Ttc8
|
tetratricopeptide repeat domain 8 |
| chr12_-_113561594 | 0.03 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr7_+_19222972 | 0.03 |
ENSMUST00000011407.8
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr11_+_75546671 | 0.03 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr8_+_61085853 | 0.02 |
ENSMUST00000161421.2
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr13_-_113800172 | 0.02 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr16_+_45430743 | 0.02 |
ENSMUST00000161347.9
ENSMUST00000023339.5 |
Gcsam
|
germinal center associated, signaling and motility |
| chr9_-_70842090 | 0.02 |
ENSMUST00000034731.10
|
Lipc
|
lipase, hepatic |
| chr7_-_90106375 | 0.02 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A |
| chr3_+_20039775 | 0.02 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr11_+_69920542 | 0.02 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr5_+_90708962 | 0.02 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr16_-_58930996 | 0.02 |
ENSMUST00000076262.4
|
Olfr193
|
olfactory receptor 193 |
| chr18_+_37822865 | 0.02 |
ENSMUST00000195112.2
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2 |
| chr14_+_54713557 | 0.01 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr15_+_78128990 | 0.01 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr16_-_26786995 | 0.01 |
ENSMUST00000231969.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr14_+_53562089 | 0.01 |
ENSMUST00000178100.3
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chrX_-_8072714 | 0.01 |
ENSMUST00000089403.10
ENSMUST00000077595.12 ENSMUST00000089402.10 ENSMUST00000082320.12 |
Porcn
|
porcupine O-acyltransferase |
| chr14_-_8551429 | 0.01 |
ENSMUST00000112652.9
|
Gm11100
|
predicted gene 11100 |
| chr19_+_34268053 | 0.01 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chrX_-_58612709 | 0.01 |
ENSMUST00000124402.2
|
Fgf13
|
fibroblast growth factor 13 |
| chr6_-_21851827 | 0.01 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr6_-_136148820 | 0.01 |
ENSMUST00000188999.3
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr7_+_27353331 | 0.01 |
ENSMUST00000008088.9
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr14_-_34225281 | 0.01 |
ENSMUST00000171343.9
|
Bmpr1a
|
bone morphogenetic protein receptor, type 1A |
| chr1_-_13062873 | 0.01 |
ENSMUST00000188454.7
|
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr18_-_39062201 | 0.01 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr10_+_129603367 | 0.01 |
ENSMUST00000060636.2
|
Olfr808
|
olfactory receptor 808 |
| chr6_-_21852508 | 0.00 |
ENSMUST00000031678.10
|
Tspan12
|
tetraspanin 12 |
| chr7_-_99276310 | 0.00 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr7_-_103778992 | 0.00 |
ENSMUST00000053743.6
|
Ubqln5
|
ubiquilin 5 |
| chr2_-_103133524 | 0.00 |
ENSMUST00000090475.10
|
Ehf
|
ets homologous factor |
| chr14_+_53007210 | 0.00 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr7_-_103721076 | 0.00 |
ENSMUST00000213184.2
ENSMUST00000213991.2 |
Olfr644
|
olfactory receptor 644 |
| chr2_-_103133503 | 0.00 |
ENSMUST00000111176.9
|
Ehf
|
ets homologous factor |
| chr6_+_89691185 | 0.00 |
ENSMUST00000075158.2
|
Vmn1r40
|
vomeronasal 1 receptor 40 |
| chr14_+_53628092 | 0.00 |
ENSMUST00000103636.4
|
Trav7-2
|
T cell receptor alpha variable 7-2 |
| chr12_+_36364136 | 0.00 |
ENSMUST00000041407.7
|
Sostdc1
|
sclerostin domain containing 1 |
| chr7_-_108493612 | 0.00 |
ENSMUST00000202706.2
ENSMUST00000084752.2 |
Olfr519
|
olfactory receptor 519 |
| chr7_-_104812698 | 0.00 |
ENSMUST00000215552.2
ENSMUST00000215744.2 ENSMUST00000217136.2 |
Olfr684
|
olfactory receptor 684 |
| chr9_-_70841881 | 0.00 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr4_-_57916283 | 0.00 |
ENSMUST00000063816.6
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr6_-_83513222 | 0.00 |
ENSMUST00000075161.12
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_-_176041498 | 0.00 |
ENSMUST00000111167.2
|
Pld5
|
phospholipase D family, member 5 |
| chr18_-_39062514 | 0.00 |
ENSMUST00000235922.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr16_-_58747023 | 0.00 |
ENSMUST00000205742.2
|
Olfr181
|
olfactory receptor 181 |
| chr14_+_53698556 | 0.00 |
ENSMUST00000181728.3
|
Trav7-4
|
T cell receptor alpha variable 7-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hivep1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 | 0.6 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.2 | 0.8 | GO:0009597 | detection of virus(GO:0009597) |
| 0.2 | 1.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.6 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.5 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.7 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.3 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.5 | GO:0071484 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 3.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 1.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.9 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.1 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.2 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.4 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.3 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 1.0 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.5 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |