Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
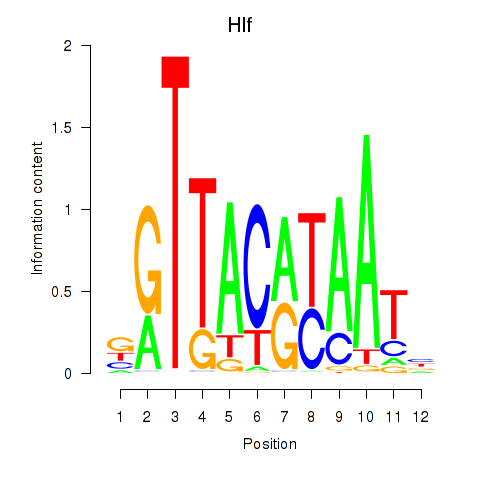
Results for Hlf
Z-value: 1.02
Transcription factors associated with Hlf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlf
|
ENSMUSG00000003949.17 | hepatic leukemia factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hlf | mm39_v1_chr11_-_90281721_90281753 | -0.86 | 6.1e-02 | Click! |
Activity profile of Hlf motif
Sorted Z-values of Hlf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_96175970 | 1.48 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr13_+_23715220 | 1.38 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr1_-_140111138 | 1.30 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 1.20 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr3_-_96170627 | 1.07 |
ENSMUST00000171473.3
|
H4c14
|
H4 clustered histone 14 |
| chr1_-_71692320 | 1.01 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr1_-_69724939 | 0.99 |
ENSMUST00000027146.9
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr18_-_36859732 | 0.55 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr10_+_23727325 | 0.45 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr16_-_36188086 | 0.41 |
ENSMUST00000096089.3
|
Cstdc5
|
cystatin domain containing 5 |
| chr11_-_115977755 | 0.40 |
ENSMUST00000074628.13
ENSMUST00000106444.4 |
Wbp2
|
WW domain binding protein 2 |
| chr7_+_27147403 | 0.37 |
ENSMUST00000037399.16
ENSMUST00000108358.8 |
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr17_+_84819260 | 0.34 |
ENSMUST00000047206.7
|
Plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr15_-_37008011 | 0.32 |
ENSMUST00000226671.2
|
Zfp706
|
zinc finger protein 706 |
| chr9_-_55190902 | 0.32 |
ENSMUST00000164721.8
|
Nrg4
|
neuregulin 4 |
| chr16_+_36097505 | 0.31 |
ENSMUST00000042097.11
|
Stfa1
|
stefin A1 |
| chr4_+_55350043 | 0.30 |
ENSMUST00000030134.9
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr11_+_69214971 | 0.29 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr1_-_69725045 | 0.29 |
ENSMUST00000190771.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr11_+_69214895 | 0.28 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr15_+_99291491 | 0.26 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_+_18618605 | 0.25 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr11_-_83540175 | 0.25 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr3_+_105778174 | 0.24 |
ENSMUST00000164730.2
ENSMUST00000010279.10 |
Adora3
Tmigd3
|
adenosine A3 receptor transmembrane and immunoglobulin domain containing 3 |
| chr11_+_62737936 | 0.24 |
ENSMUST00000150989.8
ENSMUST00000176577.2 |
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr11_+_69214883 | 0.24 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr11_+_69214789 | 0.23 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr8_+_46924074 | 0.23 |
ENSMUST00000034046.13
ENSMUST00000211644.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr7_-_140596811 | 0.22 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr4_+_40269563 | 0.22 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr11_+_118913788 | 0.21 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr13_-_74956640 | 0.21 |
ENSMUST00000231578.2
|
Cast
|
calpastatin |
| chr10_-_111833138 | 0.20 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr13_-_74956924 | 0.20 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr9_+_64086553 | 0.19 |
ENSMUST00000034965.8
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr11_+_62737887 | 0.19 |
ENSMUST00000036085.11
|
Fbxw10
|
F-box and WD-40 domain protein 10 |
| chr16_+_36004452 | 0.18 |
ENSMUST00000114858.2
|
Cstdc4
|
cystatin domain containing 4 |
| chr19_-_24938909 | 0.18 |
ENSMUST00000025815.10
|
Cbwd1
|
COBW domain containing 1 |
| chr4_-_41774097 | 0.18 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr3_+_30847024 | 0.17 |
ENSMUST00000029256.9
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr7_-_5152669 | 0.17 |
ENSMUST00000236378.2
|
Vmn1r55
|
vomeronasal 1 receptor 55 |
| chr15_+_101152078 | 0.17 |
ENSMUST00000228985.2
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_-_44145830 | 0.17 |
ENSMUST00000118515.9
ENSMUST00000138328.3 ENSMUST00000239015.2 ENSMUST00000118808.9 |
Emc10
|
ER membrane protein complex subunit 10 |
| chr16_-_75706161 | 0.16 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr18_+_23937019 | 0.16 |
ENSMUST00000025127.5
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_+_108216233 | 0.16 |
ENSMUST00000082429.8
|
Gpx1
|
glutathione peroxidase 1 |
| chr16_+_51870001 | 0.15 |
ENSMUST00000227756.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr9_-_71070506 | 0.15 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr5_+_24305577 | 0.14 |
ENSMUST00000030841.10
ENSMUST00000163409.5 |
Klhl7
|
kelch-like 7 |
| chr2_+_155118217 | 0.14 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr10_+_57521930 | 0.14 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr2_+_85820648 | 0.14 |
ENSMUST00000216807.2
|
Olfr1031
|
olfactory receptor 1031 |
| chr11_+_69011230 | 0.14 |
ENSMUST00000024543.3
|
Hes7
|
hes family bHLH transcription factor 7 |
| chr19_-_3979723 | 0.14 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr19_-_7183596 | 0.13 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr19_-_7183626 | 0.13 |
ENSMUST00000025679.11
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr10_+_57521958 | 0.13 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr18_+_35904541 | 0.13 |
ENSMUST00000170693.9
ENSMUST00000237984.2 ENSMUST00000167406.2 |
Ube2d2a
|
ubiquitin-conjugating enzyme E2D 2A |
| chr4_-_147953088 | 0.13 |
ENSMUST00000030886.15
ENSMUST00000172710.8 |
Miip
|
migration and invasion inhibitory protein |
| chr5_-_124327776 | 0.13 |
ENSMUST00000159677.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr4_-_133972890 | 0.12 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr1_-_59709898 | 0.12 |
ENSMUST00000185265.2
|
Sumo1
|
small ubiquitin-like modifier 1 |
| chr19_-_40502117 | 0.12 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_+_43741513 | 0.12 |
ENSMUST00000112047.10
|
Cutc
|
cutC copper transporter |
| chr2_-_104324035 | 0.12 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr3_-_36107661 | 0.12 |
ENSMUST00000166644.8
ENSMUST00000200469.5 |
Ccdc144b
|
coiled-coil domain containing 144B |
| chr10_-_128361731 | 0.12 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr11_-_97673203 | 0.12 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr19_+_43741431 | 0.12 |
ENSMUST00000026199.14
|
Cutc
|
cutC copper transporter |
| chr3_-_142101339 | 0.12 |
ENSMUST00000198381.5
ENSMUST00000090134.12 ENSMUST00000196908.5 |
Pdlim5
|
PDZ and LIM domain 5 |
| chr17_+_79922329 | 0.12 |
ENSMUST00000040368.3
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_-_120439764 | 0.11 |
ENSMUST00000102496.8
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr17_-_48002328 | 0.11 |
ENSMUST00000152214.4
ENSMUST00000113299.6 |
Prickle4
|
prickle planar cell polarity protein 4 |
| chr2_+_85545763 | 0.11 |
ENSMUST00000216443.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr2_+_172235820 | 0.11 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr16_+_19916292 | 0.11 |
ENSMUST00000023509.5
ENSMUST00000232088.2 ENSMUST00000231842.2 |
Klhl24
|
kelch-like 24 |
| chr3_-_127346882 | 0.10 |
ENSMUST00000197668.2
ENSMUST00000029588.10 |
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr14_+_51181956 | 0.10 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr3_+_87813624 | 0.10 |
ENSMUST00000005017.15
|
Hdgf
|
heparin binding growth factor |
| chr9_+_59564482 | 0.10 |
ENSMUST00000216620.2
ENSMUST00000217038.2 |
Pkm
|
pyruvate kinase, muscle |
| chr6_+_124785621 | 0.10 |
ENSMUST00000047760.10
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chr9_-_123546605 | 0.10 |
ENSMUST00000163207.2
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr7_-_5416395 | 0.10 |
ENSMUST00000228728.2
ENSMUST00000108569.4 |
Vmn1r58
|
vomeronasal 1 receptor 58 |
| chr17_+_47604995 | 0.10 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr6_-_115014777 | 0.10 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr7_+_102954855 | 0.10 |
ENSMUST00000214577.2
|
Olfr596
|
olfactory receptor 596 |
| chr11_+_98828495 | 0.09 |
ENSMUST00000107475.9
ENSMUST00000068133.10 |
Rara
|
retinoic acid receptor, alpha |
| chr16_+_3702523 | 0.09 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr8_-_91860576 | 0.09 |
ENSMUST00000120213.9
ENSMUST00000109609.9 |
Aktip
|
AKT interacting protein |
| chr5_+_77122530 | 0.09 |
ENSMUST00000101087.10
ENSMUST00000120550.2 |
Srp72
|
signal recognition particle 72 |
| chr9_+_115738523 | 0.09 |
ENSMUST00000119291.8
ENSMUST00000069651.13 |
Gadl1
|
glutamate decarboxylase-like 1 |
| chr14_+_51193449 | 0.09 |
ENSMUST00000095925.5
|
Pnp2
|
purine-nucleoside phosphorylase 2 |
| chr1_-_92401459 | 0.09 |
ENSMUST00000185251.2
ENSMUST00000027478.7 |
Ndufa10
|
NADH:ubiquinone oxidoreductase subunit A10 |
| chr14_+_53994813 | 0.09 |
ENSMUST00000180380.3
|
Trav13-4-dv7
|
T cell receptor alpha variable 13-4-DV7 |
| chr9_+_108216466 | 0.09 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr10_+_127256736 | 0.09 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr16_-_36154692 | 0.09 |
ENSMUST00000114850.3
|
Cstdc6
|
cystatin domain containing 6 |
| chr15_-_37007623 | 0.09 |
ENSMUST00000078976.9
|
Zfp706
|
zinc finger protein 706 |
| chr16_-_16345197 | 0.08 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr10_-_125225298 | 0.08 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr11_-_6394352 | 0.08 |
ENSMUST00000093346.6
|
H2az2
|
H2A.Z histone variant 2 |
| chr2_+_181139016 | 0.08 |
ENSMUST00000108799.10
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr4_-_120874376 | 0.08 |
ENSMUST00000043200.8
|
Smap2
|
small ArfGAP 2 |
| chr2_-_89444463 | 0.08 |
ENSMUST00000111532.4
ENSMUST00000216424.2 |
Olfr1247
|
olfactory receptor 1247 |
| chr8_-_123202774 | 0.07 |
ENSMUST00000014614.4
|
Rnf166
|
ring finger protein 166 |
| chr4_-_108690805 | 0.07 |
ENSMUST00000102742.11
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chrX_-_8042129 | 0.07 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr9_-_121534556 | 0.07 |
ENSMUST00000154978.3
|
Sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr9_-_123546642 | 0.07 |
ENSMUST00000165754.8
ENSMUST00000026274.14 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr7_+_27147475 | 0.07 |
ENSMUST00000133750.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr11_+_4014841 | 0.07 |
ENSMUST00000068322.7
|
Sec14l3
|
SEC14-like lipid binding 3 |
| chrX_+_10583629 | 0.07 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr10_+_94412116 | 0.07 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr14_-_86986541 | 0.07 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr8_+_95113066 | 0.07 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr7_+_45271229 | 0.07 |
ENSMUST00000033100.5
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr7_+_67297152 | 0.07 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_-_86646118 | 0.07 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr11_-_66059330 | 0.07 |
ENSMUST00000080665.10
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr10_+_127256993 | 0.07 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr12_-_65219328 | 0.07 |
ENSMUST00000124201.2
ENSMUST00000052201.9 ENSMUST00000222244.2 ENSMUST00000221296.2 |
Mis18bp1
|
MIS18 binding protein 1 |
| chrX_-_123045649 | 0.07 |
ENSMUST00000094491.5
|
Vmn2r121
|
vomeronasal 2, receptor 121 |
| chr17_-_83821716 | 0.07 |
ENSMUST00000025095.9
ENSMUST00000167741.9 |
Cox7a2l
|
cytochrome c oxidase subunit 7A2 like |
| chr13_+_108996606 | 0.06 |
ENSMUST00000177907.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr13_-_100787481 | 0.06 |
ENSMUST00000177848.3
ENSMUST00000022136.14 |
Rad17
|
RAD17 checkpoint clamp loader component |
| chr12_+_98737274 | 0.06 |
ENSMUST00000021399.9
|
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr4_+_130001349 | 0.06 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr2_+_109111083 | 0.06 |
ENSMUST00000028527.8
|
Kif18a
|
kinesin family member 18A |
| chr2_+_112096154 | 0.06 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr2_-_87868043 | 0.06 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73 |
| chr2_+_172314433 | 0.06 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr5_-_139446260 | 0.06 |
ENSMUST00000198474.2
|
3110082I17Rik
|
RIKEN cDNA 3110082I17 gene |
| chr11_-_69576363 | 0.05 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr16_+_43330630 | 0.05 |
ENSMUST00000114695.3
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_10505113 | 0.05 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chr14_-_20026761 | 0.05 |
ENSMUST00000161247.2
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_+_103684652 | 0.05 |
ENSMUST00000213214.2
|
Olfr641
|
olfactory receptor 641 |
| chr19_+_28941292 | 0.05 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr2_-_90735171 | 0.05 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr6_-_129599645 | 0.05 |
ENSMUST00000032252.8
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr6_-_55658242 | 0.05 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr19_-_33728759 | 0.05 |
ENSMUST00000147153.4
|
Lipo2
|
lipase, member O2 |
| chr6_+_54658609 | 0.05 |
ENSMUST00000190641.7
ENSMUST00000187701.2 |
Mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_108306267 | 0.05 |
ENSMUST00000147251.2
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr9_+_108216433 | 0.05 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr7_+_44145987 | 0.05 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chrX_-_50235283 | 0.05 |
ENSMUST00000041495.14
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr16_+_3702493 | 0.05 |
ENSMUST00000176233.2
|
Gm20695
|
predicted gene 20695 |
| chr12_+_37291632 | 0.04 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr15_+_44482944 | 0.04 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr5_-_139446244 | 0.04 |
ENSMUST00000066052.12
|
3110082I17Rik
|
RIKEN cDNA 3110082I17 gene |
| chr14_+_53599724 | 0.04 |
ENSMUST00000196105.2
|
Trav13n-4
|
T cell receptor alpha variable 13N-4 |
| chr19_-_33495180 | 0.04 |
ENSMUST00000143522.9
|
Lipo4
|
lipase, member O4 |
| chr15_-_27788693 | 0.04 |
ENSMUST00000226287.2
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr19_+_56814700 | 0.04 |
ENSMUST00000078723.11
ENSMUST00000121249.8 |
Tdrd1
|
tudor domain containing 1 |
| chr2_+_24235300 | 0.04 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr18_+_37617848 | 0.04 |
ENSMUST00000053856.6
|
Pcdhb17
|
protocadherin beta 17 |
| chrX_+_93278203 | 0.04 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15 |
| chr7_+_44146012 | 0.04 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr8_-_118398264 | 0.04 |
ENSMUST00000037955.14
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr7_-_114235506 | 0.04 |
ENSMUST00000205714.2
ENSMUST00000206853.2 ENSMUST00000205933.2 ENSMUST00000206156.2 ENSMUST00000032907.9 ENSMUST00000032906.11 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr16_+_3702604 | 0.04 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr10_+_21253190 | 0.04 |
ENSMUST00000042699.14
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr8_-_13612397 | 0.04 |
ENSMUST00000187391.7
ENSMUST00000134023.9 ENSMUST00000151400.10 |
1700029H14Rik
|
RIKEN cDNA 1700029H14 gene |
| chr3_+_103740056 | 0.04 |
ENSMUST00000106822.2
|
Bcl2l15
|
BCLl2-like 15 |
| chr6_-_128401832 | 0.04 |
ENSMUST00000203374.3
ENSMUST00000142615.4 ENSMUST00000001559.11 |
Itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr5_-_148931957 | 0.04 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
| chr2_-_120439826 | 0.04 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr2_+_124994425 | 0.04 |
ENSMUST00000110494.9
ENSMUST00000110495.3 ENSMUST00000028630.9 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr4_+_42735912 | 0.04 |
ENSMUST00000107984.2
|
4930578G10Rik
|
RIKEN cDNA 4930578G10 gene |
| chr2_+_103858168 | 0.04 |
ENSMUST00000111135.8
ENSMUST00000111136.8 ENSMUST00000102565.4 |
Fbxo3
|
F-box protein 3 |
| chr15_+_34453432 | 0.04 |
ENSMUST00000060894.9
|
Erich5
|
glutamate rich 5 |
| chr13_-_43632368 | 0.04 |
ENSMUST00000222651.2
|
Ranbp9
|
RAN binding protein 9 |
| chr17_-_45903494 | 0.04 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr13_+_49658249 | 0.04 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chrY_-_70026134 | 0.03 |
ENSMUST00000188554.7
ENSMUST00000191151.2 |
Gm28079
|
predicted gene 28079 |
| chr8_-_126625029 | 0.03 |
ENSMUST00000047239.13
ENSMUST00000131127.3 |
Pcnx2
|
pecanex homolog 2 |
| chr14_+_53310220 | 0.03 |
ENSMUST00000196079.2
|
Trav13d-4
|
T cell receptor alpha variable 13D-4 |
| chr9_+_32283779 | 0.03 |
ENSMUST00000047334.10
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr2_+_18703797 | 0.03 |
ENSMUST00000095132.10
|
Spag6
|
sperm associated antigen 6 |
| chr7_-_45343785 | 0.03 |
ENSMUST00000040636.9
|
Sec1
|
secretory blood group 1 |
| chr4_+_130253925 | 0.03 |
ENSMUST00000105994.4
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr11_-_58425662 | 0.03 |
ENSMUST00000213188.3
|
Olfr330
|
olfactory receptor 330 |
| chr17_-_45047521 | 0.03 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr17_+_31810693 | 0.03 |
ENSMUST00000236493.2
ENSMUST00000237038.2 ENSMUST00000237748.2 ENSMUST00000235255.2 |
Pknox1
|
Pbx/knotted 1 homeobox |
| chr2_-_87798643 | 0.03 |
ENSMUST00000099841.4
|
Olfr1157
|
olfactory receptor 1157 |
| chr4_+_156683398 | 0.03 |
ENSMUST00000074107.13
ENSMUST00000096792.8 |
Vmn2r129
|
vomeronasal 2, receptor 129 |
| chr19_-_34724689 | 0.03 |
ENSMUST00000009522.4
|
Slc16a12
|
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
| chr11_-_73290321 | 0.03 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr6_-_50359797 | 0.03 |
ENSMUST00000114468.9
|
Osbpl3
|
oxysterol binding protein-like 3 |
| chr2_+_164721277 | 0.03 |
ENSMUST00000041643.5
|
Pcif1
|
phosphorylated CTD interacting factor 1 |
| chr2_+_146854916 | 0.03 |
ENSMUST00000028921.6
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr19_+_31846154 | 0.03 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr16_-_4607751 | 0.03 |
ENSMUST00000117713.8
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr19_-_50667079 | 0.03 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr19_+_34268053 | 0.03 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr13_-_31131742 | 0.03 |
ENSMUST00000102943.2
|
Hus1b
|
HUS1 checkpoint clamp component B |
| chr18_-_35631914 | 0.03 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr11_+_110888313 | 0.03 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr8_-_23143422 | 0.03 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hlf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.3 | 2.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.6 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.3 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.1 | 0.4 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.2 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.4 | GO:0007343 | egg activation(GO:0007343) negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.0 | 0.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.0 | GO:0097037 | heme export(GO:0097037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.4 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.1 | 1.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |