Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
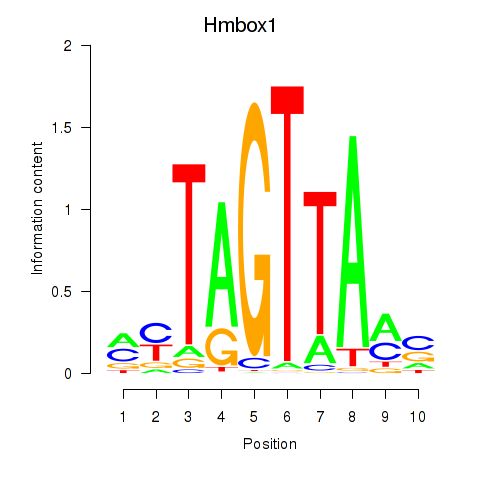
Results for Hmbox1
Z-value: 1.02
Transcription factors associated with Hmbox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmbox1
|
ENSMUSG00000021972.15 | homeobox containing 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmbox1 | mm39_v1_chr14_-_65187076_65187122 | 0.39 | 5.2e-01 | Click! |
Activity profile of Hmbox1 motif
Sorted Z-values of Hmbox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_22220000 | 1.93 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr10_+_129072073 | 0.95 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774 |
| chr16_+_3408906 | 0.81 |
ENSMUST00000216259.2
|
Olfr161
|
olfactory receptor 161 |
| chr13_+_49658249 | 0.81 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr1_+_163979384 | 0.79 |
ENSMUST00000086040.6
|
F5
|
coagulation factor V |
| chr13_+_47347301 | 0.66 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr11_-_94932158 | 0.64 |
ENSMUST00000038431.8
|
Pdk2
|
pyruvate dehydrogenase kinase, isoenzyme 2 |
| chr4_+_100336003 | 0.58 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr10_+_129084281 | 0.55 |
ENSMUST00000214109.2
|
Olfr775
|
olfactory receptor 775 |
| chr13_+_23758555 | 0.51 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr16_-_48592372 | 0.47 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr19_+_13339600 | 0.46 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chr2_+_32253016 | 0.46 |
ENSMUST00000132028.8
ENSMUST00000136079.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr10_-_95159933 | 0.45 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chrM_+_5319 | 0.44 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr10_+_129403524 | 0.42 |
ENSMUST00000204820.3
|
Olfr794
|
olfactory receptor 794 |
| chr1_-_31261678 | 0.40 |
ENSMUST00000187892.8
ENSMUST00000233331.2 |
4931428L18Rik
|
RIKEN cDNA 4931428L18 gene |
| chr7_-_85525474 | 0.40 |
ENSMUST00000077478.8
ENSMUST00000233729.2 ENSMUST00000233079.2 ENSMUST00000233315.2 ENSMUST00000233115.2 ENSMUST00000233748.2 ENSMUST00000232965.2 ENSMUST00000232897.2 |
Vmn2r73
|
vomeronasal 2, receptor 73 |
| chr13_+_56757389 | 0.38 |
ENSMUST00000045173.10
|
Tgfbi
|
transforming growth factor, beta induced |
| chr1_+_59296065 | 0.38 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr15_+_54975713 | 0.36 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr18_+_37453427 | 0.33 |
ENSMUST00000078271.4
|
Pcdhb5
|
protocadherin beta 5 |
| chr7_-_126101555 | 0.33 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like |
| chr9_-_39161290 | 0.33 |
ENSMUST00000213472.2
|
Olfr1537
|
olfactory receptor 1537 |
| chr11_+_93886906 | 0.32 |
ENSMUST00000041956.14
|
Spag9
|
sperm associated antigen 9 |
| chr10_-_12743915 | 0.32 |
ENSMUST00000219584.2
|
Utrn
|
utrophin |
| chr5_-_136594286 | 0.31 |
ENSMUST00000176172.8
|
Cux1
|
cut-like homeobox 1 |
| chr18_+_35695736 | 0.31 |
ENSMUST00000235851.2
ENSMUST00000235581.2 |
Matr3
|
matrin 3 |
| chr12_-_69939931 | 0.30 |
ENSMUST00000049239.8
ENSMUST00000110570.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr15_+_54975814 | 0.29 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr14_-_55909314 | 0.29 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr12_-_69939762 | 0.29 |
ENSMUST00000110567.8
ENSMUST00000171211.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_+_32253204 | 0.29 |
ENSMUST00000048964.14
ENSMUST00000113332.8 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr4_+_116415251 | 0.28 |
ENSMUST00000106475.2
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr12_+_69939879 | 0.26 |
ENSMUST00000021466.10
|
Atl1
|
atlastin GTPase 1 |
| chr6_+_123100382 | 0.26 |
ENSMUST00000032248.8
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr11_-_49005701 | 0.26 |
ENSMUST00000060398.3
ENSMUST00000215553.2 ENSMUST00000109201.2 |
Olfr1396
|
olfactory receptor 1396 |
| chr10_-_128918779 | 0.24 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr6_+_123100272 | 0.24 |
ENSMUST00000041779.13
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr12_-_65012270 | 0.24 |
ENSMUST00000222508.2
|
Klhl28
|
kelch-like 28 |
| chr1_+_173093568 | 0.21 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr2_+_32253692 | 0.20 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr3_+_28752050 | 0.19 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr8_+_45960804 | 0.19 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr16_+_36073416 | 0.19 |
ENSMUST00000063539.7
|
Csta2
|
cystatin A family member 2 |
| chr10_-_81243475 | 0.17 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr3_-_95125051 | 0.17 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr14_+_66872699 | 0.16 |
ENSMUST00000159365.8
ENSMUST00000054661.8 ENSMUST00000225182.2 ENSMUST00000159068.2 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr18_+_65831324 | 0.15 |
ENSMUST00000115097.8
ENSMUST00000117694.2 ENSMUST00000235962.2 |
Oacyl
|
O-acyltransferase like |
| chr6_+_123099615 | 0.15 |
ENSMUST00000161636.8
ENSMUST00000161365.8 |
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_-_16549394 | 0.15 |
ENSMUST00000206259.2
|
Fkrp
|
fukutin related protein |
| chr15_+_99291491 | 0.15 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_20042045 | 0.15 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chr11_+_77576981 | 0.14 |
ENSMUST00000100802.11
ENSMUST00000181023.2 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr4_+_155709275 | 0.14 |
ENSMUST00000067081.10
ENSMUST00000105600.8 ENSMUST00000105598.2 |
Cdk11b
|
cyclin-dependent kinase 11B |
| chr14_+_54032814 | 0.14 |
ENSMUST00000103671.4
|
Trav13-5
|
T cell receptor alpha variable 13-5 |
| chr9_+_18848418 | 0.14 |
ENSMUST00000218385.2
|
Olfr832
|
olfactory receptor 832 |
| chr6_-_97156032 | 0.14 |
ENSMUST00000095664.6
|
Tmf1
|
TATA element modulatory factor 1 |
| chr7_+_7174315 | 0.14 |
ENSMUST00000051435.8
|
Zfp418
|
zinc finger protein 418 |
| chr11_+_31822211 | 0.13 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr14_-_36628263 | 0.13 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr11_+_22958338 | 0.13 |
ENSMUST00000172602.9
|
Fam161a
|
family with sequence similarity 161, member A |
| chr12_-_113324852 | 0.13 |
ENSMUST00000223179.2
ENSMUST00000103423.3 |
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr13_-_73826124 | 0.13 |
ENSMUST00000022105.15
ENSMUST00000109680.10 ENSMUST00000221026.2 ENSMUST00000109679.4 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr11_-_4654303 | 0.13 |
ENSMUST00000058407.6
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr2_+_96148418 | 0.12 |
ENSMUST00000135431.8
ENSMUST00000162807.9 |
Lrrc4c
|
leucine rich repeat containing 4C |
| chr15_+_82152994 | 0.12 |
ENSMUST00000238416.3
ENSMUST00000239048.2 |
Septin3
|
septin 3 |
| chr4_-_133066594 | 0.12 |
ENSMUST00000043305.14
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr16_-_21814289 | 0.12 |
ENSMUST00000060673.8
|
Liph
|
lipase, member H |
| chr13_-_96678844 | 0.11 |
ENSMUST00000223475.2
|
Polk
|
polymerase (DNA directed), kappa |
| chr11_-_78313043 | 0.11 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr3_-_68911807 | 0.11 |
ENSMUST00000154741.8
ENSMUST00000148031.2 |
Ift80
|
intraflagellar transport 80 |
| chr19_+_32573182 | 0.10 |
ENSMUST00000235594.2
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr4_-_133066549 | 0.10 |
ENSMUST00000105906.2
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr8_-_66939146 | 0.10 |
ENSMUST00000026681.7
|
Tma16
|
translation machinery associated 16 |
| chr13_-_3943433 | 0.10 |
ENSMUST00000222504.2
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr9_+_123902143 | 0.09 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr2_-_12424212 | 0.08 |
ENSMUST00000124603.8
ENSMUST00000129993.3 ENSMUST00000028105.13 |
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr3_-_144525255 | 0.08 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr8_+_106587212 | 0.07 |
ENSMUST00000008594.9
|
Nutf2
|
nuclear transport factor 2 |
| chr4_-_148021217 | 0.07 |
ENSMUST00000019199.14
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr13_-_12535236 | 0.07 |
ENSMUST00000179308.3
|
Edaradd
|
EDAR (ectodysplasin-A receptor)-associated death domain |
| chr14_-_10185787 | 0.07 |
ENSMUST00000225871.2
|
Gm49355
|
predicted gene, 49355 |
| chr1_-_69726384 | 0.06 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr4_+_116414855 | 0.06 |
ENSMUST00000030460.15
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr18_-_10706701 | 0.06 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr4_+_149602673 | 0.06 |
ENSMUST00000030839.13
|
Ctnnbip1
|
catenin beta interacting protein 1 |
| chr11_+_11414256 | 0.06 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr6_-_121058551 | 0.06 |
ENSMUST00000077159.8
ENSMUST00000207889.2 ENSMUST00000238957.2 ENSMUST00000238920.2 ENSMUST00000238968.2 |
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr10_-_40133558 | 0.06 |
ENSMUST00000216847.2
ENSMUST00000213628.2 ENSMUST00000217537.2 ENSMUST00000019982.9 |
Gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr2_+_173561208 | 0.06 |
ENSMUST00000073081.6
|
1700010B08Rik
|
RIKEN cDNA 1700010B08 gene |
| chr1_-_171476559 | 0.06 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chr15_-_78058214 | 0.05 |
ENSMUST00000016781.8
|
Ift27
|
intraflagellar transport 27 |
| chr4_-_46602192 | 0.05 |
ENSMUST00000107756.4
|
Coro2a
|
coronin, actin binding protein 2A |
| chr19_-_3332955 | 0.05 |
ENSMUST00000119292.8
ENSMUST00000025751.11 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr19_-_28945194 | 0.04 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr3_+_144824644 | 0.04 |
ENSMUST00000199124.5
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr3_-_108797022 | 0.04 |
ENSMUST00000180063.8
ENSMUST00000053065.8 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr7_-_38227617 | 0.04 |
ENSMUST00000079759.6
|
Gm5591
|
predicted gene 5591 |
| chr8_-_106863423 | 0.04 |
ENSMUST00000146940.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr1_-_185849448 | 0.04 |
ENSMUST00000045388.8
|
Lyplal1
|
lysophospholipase-like 1 |
| chr14_-_55101505 | 0.04 |
ENSMUST00000142283.4
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr4_-_58787716 | 0.04 |
ENSMUST00000216719.2
|
Olfr267
|
olfactory receptor 267 |
| chr1_-_171476495 | 0.04 |
ENSMUST00000194791.2
ENSMUST00000192024.6 |
Slamf7
|
SLAM family member 7 |
| chr1_-_22845124 | 0.03 |
ENSMUST00000115273.10
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr19_+_8734503 | 0.03 |
ENSMUST00000183939.2
|
Nxf1
|
nuclear RNA export factor 1 |
| chr1_-_133589020 | 0.03 |
ENSMUST00000193504.6
ENSMUST00000195067.2 ENSMUST00000191896.6 ENSMUST00000194668.6 ENSMUST00000195424.6 ENSMUST00000179598.4 ENSMUST00000027736.13 |
Zc3h11a
Gm38394
Zc3h11a
|
zinc finger CCCH type containing 11A predicted gene, 38394 zinc finger CCCH type containing 11A |
| chr6_+_96092230 | 0.03 |
ENSMUST00000075080.6
|
Tafa1
|
TAFA chemokine like family member 1 |
| chr3_-_57202301 | 0.03 |
ENSMUST00000171384.8
|
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr9_-_49397508 | 0.03 |
ENSMUST00000055096.5
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr13_-_3943556 | 0.03 |
ENSMUST00000099946.6
|
Net1
|
neuroepithelial cell transforming gene 1 |
| chr10_-_128885867 | 0.03 |
ENSMUST00000216460.2
|
Olfr765
|
olfactory receptor 765 |
| chr2_+_34764408 | 0.03 |
ENSMUST00000113068.9
ENSMUST00000047447.13 |
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr1_+_24216691 | 0.03 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr7_-_130748035 | 0.02 |
ENSMUST00000070980.4
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene |
| chr13_-_73826108 | 0.02 |
ENSMUST00000222029.2
ENSMUST00000223074.2 ENSMUST00000220650.2 ENSMUST00000221987.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr12_-_81579614 | 0.02 |
ENSMUST00000169158.2
ENSMUST00000164431.2 ENSMUST00000163402.8 ENSMUST00000166664.2 ENSMUST00000164386.8 |
Synj2bp
Gm20498
|
synaptojanin 2 binding protein predicted gene 20498 |
| chr7_-_39062584 | 0.02 |
ENSMUST00000108017.2
|
Gm5114
|
predicted gene 5114 |
| chr1_+_182591425 | 0.02 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr15_+_100320028 | 0.02 |
ENSMUST00000132119.2
|
Gm5475
|
predicted gene 5475 |
| chr2_+_34764496 | 0.02 |
ENSMUST00000028228.6
|
Cutal
|
cutA divalent cation tolerance homolog-like |
| chr13_+_55359866 | 0.02 |
ENSMUST00000224693.2
|
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr2_+_119039456 | 0.02 |
ENSMUST00000102519.5
|
Zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr14_+_54198389 | 0.02 |
ENSMUST00000103678.4
|
Trdv2-2
|
T cell receptor delta variable 2-2 |
| chr7_-_109322993 | 0.02 |
ENSMUST00000106735.9
ENSMUST00000033334.5 |
BC051019
|
cDNA sequence BC051019 |
| chr1_+_21310821 | 0.02 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr17_-_13070780 | 0.02 |
ENSMUST00000162389.2
ENSMUST00000162119.8 ENSMUST00000159223.8 |
Mas1
|
MAS1 oncogene |
| chrX_-_12026594 | 0.02 |
ENSMUST00000043441.13
|
Bcor
|
BCL6 interacting corepressor |
| chr9_+_106391771 | 0.02 |
ENSMUST00000085113.5
|
Iqcf5
|
IQ motif containing F5 |
| chr14_-_63430937 | 0.02 |
ENSMUST00000225157.2
ENSMUST00000226002.2 ENSMUST00000038229.5 |
Neil2
|
nei like 2 (E. coli) |
| chr14_+_27344385 | 0.02 |
ENSMUST00000210135.2
ENSMUST00000090302.6 ENSMUST00000211087.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr6_+_33226020 | 0.01 |
ENSMUST00000052266.15
ENSMUST00000090381.11 ENSMUST00000115080.2 |
Exoc4
|
exocyst complex component 4 |
| chr3_-_123029782 | 0.01 |
ENSMUST00000106427.8
ENSMUST00000198584.2 |
Synpo2
|
synaptopodin 2 |
| chr12_-_115157739 | 0.01 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr7_+_40682143 | 0.01 |
ENSMUST00000164422.2
|
Gm4884
|
predicted gene 4884 |
| chr7_-_79036222 | 0.01 |
ENSMUST00000205638.2
ENSMUST00000206320.3 ENSMUST00000205442.2 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr10_+_4296806 | 0.01 |
ENSMUST00000216139.3
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr3_+_117368483 | 0.01 |
ENSMUST00000039564.11
ENSMUST00000238937.2 |
Plppr5
|
phospholipid phosphatase related 5 |
| chr12_-_114909863 | 0.01 |
ENSMUST00000103517.2
ENSMUST00000195417.2 |
Ighv1-43
|
immunoglobulin heavy variable V1-43 |
| chr5_-_38719025 | 0.01 |
ENSMUST00000005234.13
|
Wdr1
|
WD repeat domain 1 |
| chr8_+_46387647 | 0.01 |
ENSMUST00000095326.10
|
Ccdc110
|
coiled-coil domain containing 110 |
| chr4_-_63090355 | 0.01 |
ENSMUST00000156618.9
ENSMUST00000030042.3 |
Kif12
|
kinesin family member 12 |
| chr1_+_31261889 | 0.01 |
ENSMUST00000027230.3
|
Dnaaf6
|
dynein axonemal assembly factor 6 |
| chr10_+_112107026 | 0.01 |
ENSMUST00000219301.2
ENSMUST00000092175.4 |
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2 |
| chr13_-_35108293 | 0.01 |
ENSMUST00000223834.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr2_-_89084074 | 0.01 |
ENSMUST00000216392.2
|
Olfr1228
|
olfactory receptor 1228 |
| chr5_+_136721938 | 0.01 |
ENSMUST00000196068.5
ENSMUST00000005611.10 |
Myl10
|
myosin, light chain 10, regulatory |
| chr11_+_73673790 | 0.01 |
ENSMUST00000120081.3
ENSMUST00000215161.2 ENSMUST00000206815.2 |
Olfr390
|
olfactory receptor 390 |
| chr5_-_70999547 | 0.01 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr8_+_110806390 | 0.01 |
ENSMUST00000212754.2
ENSMUST00000058804.10 |
Zfp612
|
zinc finger protein 612 |
| chr12_-_114330574 | 0.01 |
ENSMUST00000103485.3
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr10_-_49659355 | 0.01 |
ENSMUST00000105484.10
ENSMUST00000218598.2 ENSMUST00000079751.9 ENSMUST00000218441.2 |
Grik2
|
glutamate receptor, ionotropic, kainate 2 (beta 2) |
| chr9_+_104443876 | 0.00 |
ENSMUST00000157006.8
|
Cpne4
|
copine IV |
| chr6_+_72074718 | 0.00 |
ENSMUST00000187007.3
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr14_-_79461316 | 0.00 |
ENSMUST00000040802.5
|
Zfp957
|
zinc finger protein 957 |
| chr13_+_68011442 | 0.00 |
ENSMUST00000078471.7
|
BC048507
|
cDNA sequence BC048507 |
| chr7_-_47677345 | 0.00 |
ENSMUST00000094390.3
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr3_-_108797306 | 0.00 |
ENSMUST00000102620.10
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr17_+_69746321 | 0.00 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chrX_-_110440860 | 0.00 |
ENSMUST00000128819.8
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr10_-_52071052 | 0.00 |
ENSMUST00000218452.2
|
Ros1
|
Ros1 proto-oncogene |
| chr18_-_38999755 | 0.00 |
ENSMUST00000115582.8
ENSMUST00000236060.2 |
Fgf1
|
fibroblast growth factor 1 |
| chr9_-_78275430 | 0.00 |
ENSMUST00000071991.6
|
Dppa5a
|
developmental pluripotency associated 5A |
| chr2_-_66271097 | 0.00 |
ENSMUST00000112371.9
ENSMUST00000138910.3 |
Scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr5_+_7354130 | 0.00 |
ENSMUST00000160634.2
ENSMUST00000159546.2 |
Tex47
|
testis expressed 47 |
| chr19_+_47854249 | 0.00 |
ENSMUST00000238084.2
|
Gsto2
|
glutathione S-transferase omega 2 |
| chrX_+_134934116 | 0.00 |
ENSMUST00000057625.3
|
Arxes1
|
adipocyte-related X-chromosome expressed sequence 1 |
| chr10_+_100428212 | 0.00 |
ENSMUST00000187119.7
ENSMUST00000188736.7 ENSMUST00000191336.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr2_+_85551751 | 0.00 |
ENSMUST00000055517.3
|
Olfr1009
|
olfactory receptor 1009 |
| chr8_+_45960855 | 0.00 |
ENSMUST00000141039.8
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr10_+_100428246 | 0.00 |
ENSMUST00000041162.13
ENSMUST00000190386.7 ENSMUST00000190708.7 |
1700017N19Rik
|
RIKEN cDNA 1700017N19 gene |
| chr11_+_49039086 | 0.00 |
ENSMUST00000059379.2
|
Olfr1395
|
olfactory receptor 1395 |
| chr7_-_106354591 | 0.00 |
ENSMUST00000214306.2
ENSMUST00000216255.2 |
Olfr698
|
olfactory receptor 698 |
| chr6_-_68732577 | 0.00 |
ENSMUST00000103332.2
|
Igkv4-92
|
immunoglobulin kappa variable 4-92 |
| chr4_+_114020581 | 0.00 |
ENSMUST00000079915.10
ENSMUST00000164297.8 |
Skint11
|
selection and upkeep of intraepithelial T cells 11 |
| chr11_+_67061908 | 0.00 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chrX_-_110447644 | 0.00 |
ENSMUST00000132319.8
ENSMUST00000123951.8 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr9_-_71075939 | 0.00 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr13_+_4484305 | 0.00 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr14_+_53491249 | 0.00 |
ENSMUST00000196941.2
|
Trav13n-2
|
T cell receptor alpha variable 13N-2 |
| chr5_+_88117307 | 0.00 |
ENSMUST00000007601.4
ENSMUST00000187738.2 |
2310003L06Rik
Gm28434
|
RIKEN cDNA 2310003L06 gene predicted gene 28434 |
| chr19_+_45003670 | 0.00 |
ENSMUST00000236377.2
|
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr7_+_104587735 | 0.00 |
ENSMUST00000098164.2
|
Olfr669
|
olfactory receptor 669 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmbox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.2 | 0.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:1902567 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.0 | 0.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.0 | 0.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |