Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
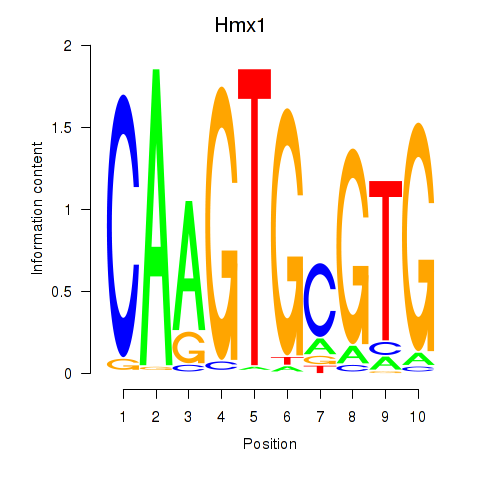
Results for Hmx1
Z-value: 1.89
Transcription factors associated with Hmx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hmx1
|
ENSMUSG00000067438.6 | H6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmx1 | mm39_v1_chr5_+_35546363_35546461 | -0.09 | 8.9e-01 | Click! |
Activity profile of Hmx1 motif
Sorted Z-values of Hmx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_127128007 | 1.72 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr9_+_108437485 | 1.39 |
ENSMUST00000081111.14
ENSMUST00000193421.2 |
Impdh2
|
inosine monophosphate dehydrogenase 2 |
| chr14_-_56339915 | 1.36 |
ENSMUST00000015583.2
|
Ctsg
|
cathepsin G |
| chr7_-_144493560 | 1.23 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr2_-_77110933 | 1.19 |
ENSMUST00000102659.2
|
Sestd1
|
SEC14 and spectrin domains 1 |
| chrX_-_133709733 | 1.16 |
ENSMUST00000035559.11
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr7_-_79882313 | 1.14 |
ENSMUST00000206084.2
ENSMUST00000205996.2 ENSMUST00000071457.12 |
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr3_-_89009153 | 1.07 |
ENSMUST00000199668.3
ENSMUST00000196709.5 |
Fdps
|
farnesyl diphosphate synthetase |
| chr17_-_34218301 | 1.04 |
ENSMUST00000235463.2
|
H2-K1
|
histocompatibility 2, K1, K region |
| chr19_+_21630887 | 0.92 |
ENSMUST00000052556.5
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr9_-_119812042 | 0.92 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr7_-_79882501 | 0.88 |
ENSMUST00000065163.15
|
Cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr3_-_89009214 | 0.85 |
ENSMUST00000081848.13
|
Fdps
|
farnesyl diphosphate synthetase |
| chr9_+_106048116 | 0.85 |
ENSMUST00000020490.13
|
Wdr82
|
WD repeat domain containing 82 |
| chr14_-_56499690 | 0.84 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr12_+_112586501 | 0.82 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr17_-_12988492 | 0.80 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr6_-_127127959 | 0.78 |
ENSMUST00000201637.2
|
Ccnd2
|
cyclin D2 |
| chr11_+_23234644 | 0.76 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr6_-_127127993 | 0.75 |
ENSMUST00000201066.2
|
Ccnd2
|
cyclin D2 |
| chr15_-_79571977 | 0.71 |
ENSMUST00000023061.7
|
Josd1
|
Josephin domain containing 1 |
| chr2_+_174169351 | 0.71 |
ENSMUST00000124935.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr14_-_79539063 | 0.70 |
ENSMUST00000022595.8
|
Rgcc
|
regulator of cell cycle |
| chr6_+_137712076 | 0.70 |
ENSMUST00000064910.7
|
Strap
|
serine/threonine kinase receptor associated protein |
| chr12_-_112893382 | 0.70 |
ENSMUST00000075827.5
|
Jag2
|
jagged 2 |
| chr14_+_55120875 | 0.70 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr12_+_112586465 | 0.70 |
ENSMUST00000021726.8
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr16_-_17906886 | 0.68 |
ENSMUST00000132241.2
ENSMUST00000139861.2 ENSMUST00000003620.13 |
Prodh
|
proline dehydrogenase |
| chr4_+_129407374 | 0.62 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr9_-_21223551 | 0.60 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr1_+_52047368 | 0.59 |
ENSMUST00000027277.7
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr7_-_119319965 | 0.58 |
ENSMUST00000033236.9
|
Thumpd1
|
THUMP domain containing 1 |
| chr13_-_95511837 | 0.58 |
ENSMUST00000022189.9
|
Aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr4_+_134195631 | 0.57 |
ENSMUST00000030636.11
ENSMUST00000127279.8 ENSMUST00000105867.8 |
Stmn1
|
stathmin 1 |
| chr14_+_54924439 | 0.57 |
ENSMUST00000227269.2
|
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr9_+_32607301 | 0.55 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr11_-_48836975 | 0.55 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr2_+_162916551 | 0.55 |
ENSMUST00000142729.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr7_-_125968653 | 0.53 |
ENSMUST00000205642.2
ENSMUST00000032997.8 ENSMUST00000206793.2 |
Lat
|
linker for activation of T cells |
| chr13_-_32522548 | 0.52 |
ENSMUST00000041859.9
|
Gmds
|
GDP-mannose 4, 6-dehydratase |
| chr15_-_31531122 | 0.51 |
ENSMUST00000090227.6
|
Marchf6
|
membrane associated ring-CH-type finger 6 |
| chr13_+_45660905 | 0.51 |
ENSMUST00000000260.13
|
Gmpr
|
guanosine monophosphate reductase |
| chr15_+_8138805 | 0.50 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chrX_+_49930311 | 0.50 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr16_+_49519561 | 0.49 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr2_-_93988229 | 0.49 |
ENSMUST00000028619.5
|
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr3_-_107992662 | 0.49 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr15_+_101982208 | 0.48 |
ENSMUST00000169681.3
ENSMUST00000229400.2 |
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr17_-_34109513 | 0.46 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr6_-_40413056 | 0.46 |
ENSMUST00000039008.10
ENSMUST00000101492.10 |
Dennd11
|
DENN domain containing 11 |
| chr5_-_134643805 | 0.46 |
ENSMUST00000202085.2
ENSMUST00000036362.13 ENSMUST00000077636.8 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr3_-_89325594 | 0.45 |
ENSMUST00000029679.4
|
Cks1b
|
CDC28 protein kinase 1b |
| chr17_+_35413415 | 0.45 |
ENSMUST00000025262.6
ENSMUST00000173600.2 |
Ltb
|
lymphotoxin B |
| chr11_-_102210568 | 0.44 |
ENSMUST00000173870.8
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr2_+_28403255 | 0.44 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr15_-_79718423 | 0.44 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr11_-_102209767 | 0.43 |
ENSMUST00000174302.8
ENSMUST00000178839.8 ENSMUST00000006754.14 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr6_-_67512768 | 0.43 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr15_+_102234802 | 0.43 |
ENSMUST00000169637.8
|
Pfdn5
|
prefoldin 5 |
| chr1_-_156546600 | 0.42 |
ENSMUST00000122424.8
ENSMUST00000086153.8 |
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
| chr8_-_25506756 | 0.42 |
ENSMUST00000084032.6
ENSMUST00000207132.2 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr19_+_21630540 | 0.42 |
ENSMUST00000235332.2
|
Abhd17b
|
abhydrolase domain containing 17B |
| chr11_-_120515799 | 0.42 |
ENSMUST00000106183.3
ENSMUST00000080202.12 |
Sirt7
|
sirtuin 7 |
| chr4_+_116078830 | 0.42 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr10_-_30494333 | 0.41 |
ENSMUST00000019925.7
|
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr15_+_44482944 | 0.41 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr12_-_111679344 | 0.41 |
ENSMUST00000160576.2
|
Bag5
|
BCL2-associated athanogene 5 |
| chr7_+_141061923 | 0.40 |
ENSMUST00000239217.2
|
Tspan4
|
tetraspanin 4 |
| chr5_-_31448370 | 0.40 |
ENSMUST00000041565.11
ENSMUST00000201809.2 |
Ift172
|
intraflagellar transport 172 |
| chr6_+_108637577 | 0.40 |
ENSMUST00000032194.11
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr13_+_91609169 | 0.40 |
ENSMUST00000004094.15
ENSMUST00000042122.15 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr18_+_38088597 | 0.40 |
ENSMUST00000070709.9
ENSMUST00000177058.8 ENSMUST00000169360.9 ENSMUST00000163591.9 ENSMUST00000091932.12 |
Rell2
|
RELT-like 2 |
| chr11_-_97520511 | 0.40 |
ENSMUST00000052281.6
|
Epop
|
elongin BC and polycomb repressive complex 2 associated protein |
| chr15_+_88703786 | 0.40 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr2_+_130116357 | 0.39 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr13_-_99481160 | 0.39 |
ENSMUST00000022153.8
|
Ptcd2
|
pentatricopeptide repeat domain 2 |
| chr14_-_56812839 | 0.39 |
ENSMUST00000225951.2
|
Cenpj
|
centromere protein J |
| chr5_+_100666278 | 0.38 |
ENSMUST00000151414.8
|
Cops4
|
COP9 signalosome subunit 4 |
| chr13_+_91609264 | 0.38 |
ENSMUST00000231481.2
|
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr8_+_124380639 | 0.38 |
ENSMUST00000045487.4
|
Rhou
|
ras homolog family member U |
| chr2_+_167922924 | 0.38 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr15_+_78312764 | 0.38 |
ENSMUST00000162517.8
ENSMUST00000166142.10 ENSMUST00000089414.11 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr4_-_98271469 | 0.38 |
ENSMUST00000143116.2
ENSMUST00000030292.12 ENSMUST00000102793.11 |
Tm2d1
|
TM2 domain containing 1 |
| chrX_-_73436609 | 0.37 |
ENSMUST00000015427.13
|
Fam3a
|
family with sequence similarity 3, member A |
| chr2_+_26279345 | 0.37 |
ENSMUST00000076431.13
ENSMUST00000114093.2 |
Pmpca
|
peptidase (mitochondrial processing) alpha |
| chr8_+_53964721 | 0.37 |
ENSMUST00000211424.2
ENSMUST00000033920.6 |
Aga
|
aspartylglucosaminidase |
| chr11_-_115258508 | 0.37 |
ENSMUST00000044152.13
ENSMUST00000106542.9 |
Hid1
|
HID1 domain containing |
| chr2_+_129040677 | 0.37 |
ENSMUST00000028880.10
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr16_+_18695787 | 0.36 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chr7_+_48438751 | 0.35 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chrX_-_73416824 | 0.35 |
ENSMUST00000178691.2
ENSMUST00000114146.8 |
Ubl4a
Slc10a3
|
ubiquitin-like 4A solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr4_-_107928567 | 0.35 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr13_+_73752125 | 0.35 |
ENSMUST00000022102.9
|
Clptm1l
|
CLPTM1-like |
| chr11_+_5470968 | 0.34 |
ENSMUST00000239150.2
|
Xbp1
|
X-box binding protein 1 |
| chr7_-_125760164 | 0.34 |
ENSMUST00000164741.2
|
Xpo6
|
exportin 6 |
| chr6_+_34453142 | 0.33 |
ENSMUST00000045372.6
ENSMUST00000138668.2 ENSMUST00000139067.2 |
Bpgm
|
2,3-bisphosphoglycerate mutase |
| chr4_+_115595610 | 0.33 |
ENSMUST00000106524.4
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr19_+_8713156 | 0.33 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr6_-_127086480 | 0.33 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr1_-_39616369 | 0.33 |
ENSMUST00000195705.2
|
Rnf149
|
ring finger protein 149 |
| chr16_-_95883714 | 0.33 |
ENSMUST00000113827.4
|
Brwd1
|
bromodomain and WD repeat domain containing 1 |
| chrX_-_73436709 | 0.32 |
ENSMUST00000114142.8
ENSMUST00000114139.8 |
Fam3a
|
family with sequence similarity 3, member A |
| chr17_+_36290743 | 0.31 |
ENSMUST00000087200.4
|
Gnl1
|
guanine nucleotide binding protein-like 1 |
| chr1_+_61017057 | 0.31 |
ENSMUST00000027162.12
ENSMUST00000102827.4 |
Icos
|
inducible T cell co-stimulator |
| chr13_-_38842967 | 0.31 |
ENSMUST00000001757.9
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr12_-_26465253 | 0.31 |
ENSMUST00000020971.14
|
Rnf144a
|
ring finger protein 144A |
| chr9_+_45817795 | 0.31 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr8_+_108162985 | 0.31 |
ENSMUST00000166615.3
ENSMUST00000213097.2 ENSMUST00000212205.2 |
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr14_-_47805861 | 0.30 |
ENSMUST00000228784.2
ENSMUST00000042988.7 |
Atg14
|
autophagy related 14 |
| chr15_+_44482545 | 0.30 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr15_+_78312851 | 0.29 |
ENSMUST00000159771.8
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr5_+_100666175 | 0.29 |
ENSMUST00000045993.15
|
Cops4
|
COP9 signalosome subunit 4 |
| chr12_+_70984631 | 0.29 |
ENSMUST00000021479.6
|
Actr10
|
ARP10 actin-related protein 10 |
| chr15_+_103411689 | 0.29 |
ENSMUST00000226493.2
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr11_+_109541747 | 0.28 |
ENSMUST00000049527.7
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chrX_+_20291927 | 0.28 |
ENSMUST00000115384.9
|
Jade3
|
jade family PHD finger 3 |
| chr11_-_109364424 | 0.27 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr1_+_135656885 | 0.27 |
ENSMUST00000027677.8
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr11_+_88864753 | 0.27 |
ENSMUST00000036649.8
|
Coil
|
coilin |
| chr12_-_56392646 | 0.27 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr15_-_79025387 | 0.27 |
ENSMUST00000187550.7
ENSMUST00000188562.7 ENSMUST00000190509.7 ENSMUST00000190730.7 ENSMUST00000190959.7 ENSMUST00000169604.8 ENSMUST00000186053.7 |
1700088E04Rik
|
RIKEN cDNA 1700088E04 gene |
| chrX_+_10583629 | 0.27 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr9_-_44646487 | 0.26 |
ENSMUST00000034611.15
|
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr17_-_27352593 | 0.26 |
ENSMUST00000118613.8
|
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chrX_-_73289970 | 0.25 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr4_+_129181407 | 0.25 |
ENSMUST00000102599.4
|
Sync
|
syncoilin |
| chr7_+_96600756 | 0.25 |
ENSMUST00000107159.3
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr4_+_109533753 | 0.25 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1 |
| chr2_-_39116044 | 0.24 |
ENSMUST00000204368.2
|
Ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr8_-_25506916 | 0.24 |
ENSMUST00000084035.12
ENSMUST00000208247.3 |
Adam9
|
a disintegrin and metallopeptidase domain 9 (meltrin gamma) |
| chr7_+_96600712 | 0.23 |
ENSMUST00000044466.12
|
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chrX_-_73416869 | 0.23 |
ENSMUST00000073067.11
ENSMUST00000037967.6 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr17_-_36291087 | 0.23 |
ENSMUST00000055454.14
|
Prr3
|
proline-rich polypeptide 3 |
| chr4_-_94444975 | 0.23 |
ENSMUST00000030313.9
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr2_-_25162347 | 0.22 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1 |
| chrX_+_158039107 | 0.22 |
ENSMUST00000148570.8
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr11_-_46203047 | 0.22 |
ENSMUST00000129474.2
ENSMUST00000093166.11 ENSMUST00000165599.9 |
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr2_-_34716083 | 0.22 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr17_+_87270707 | 0.22 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr2_+_146854916 | 0.22 |
ENSMUST00000028921.6
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr19_-_41969558 | 0.21 |
ENSMUST00000026168.9
ENSMUST00000171561.8 |
Mms19
|
MMS19 cytosolic iron-sulfur assembly component |
| chr7_-_101570393 | 0.21 |
ENSMUST00000106965.8
ENSMUST00000106968.8 ENSMUST00000106967.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr19_-_55087849 | 0.21 |
ENSMUST00000061856.6
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chrX_-_73290140 | 0.21 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr11_+_95304903 | 0.21 |
ENSMUST00000107724.9
ENSMUST00000150884.8 ENSMUST00000107722.8 ENSMUST00000127713.2 |
Spop
|
speckle-type BTB/POZ protein |
| chr16_-_97564910 | 0.20 |
ENSMUST00000019386.10
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr14_+_25842597 | 0.20 |
ENSMUST00000112364.8
|
Anxa11
|
annexin A11 |
| chrX_-_73436293 | 0.19 |
ENSMUST00000114138.8
|
Fam3a
|
family with sequence similarity 3, member A |
| chr13_+_99481283 | 0.19 |
ENSMUST00000052249.7
ENSMUST00000224660.3 |
Mrps27
|
mitochondrial ribosomal protein S27 |
| chr4_+_135648041 | 0.19 |
ENSMUST00000030434.5
|
Fuca1
|
fucosidase, alpha-L- 1, tissue |
| chr7_-_30614249 | 0.19 |
ENSMUST00000190950.7
ENSMUST00000187137.7 ENSMUST00000190638.7 |
Mag
|
myelin-associated glycoprotein |
| chr15_+_44482667 | 0.19 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr2_+_28403084 | 0.19 |
ENSMUST00000135803.8
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr7_+_132212349 | 0.18 |
ENSMUST00000033241.6
ENSMUST00000106170.8 |
Lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr17_-_27352876 | 0.18 |
ENSMUST00000119227.3
ENSMUST00000025045.15 |
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr1_+_37911272 | 0.18 |
ENSMUST00000041621.5
|
Lipt1
|
lipoyltransferase 1 |
| chr1_-_181669891 | 0.18 |
ENSMUST00000193028.2
ENSMUST00000191878.6 ENSMUST00000005003.12 |
Lbr
|
lamin B receptor |
| chr3_+_108191398 | 0.18 |
ENSMUST00000135636.6
ENSMUST00000102632.7 |
Sort1
|
sortilin 1 |
| chrX_-_101687813 | 0.18 |
ENSMUST00000052012.14
ENSMUST00000043596.12 ENSMUST00000119229.8 ENSMUST00000122022.8 ENSMUST00000120270.8 ENSMUST00000113611.3 |
Phka1
|
phosphorylase kinase alpha 1 |
| chr7_+_118311740 | 0.18 |
ENSMUST00000106557.8
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr2_-_26127360 | 0.18 |
ENSMUST00000036187.9
|
Qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr10_+_20024203 | 0.17 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr3_-_51184730 | 0.16 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr18_+_24737009 | 0.16 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr18_+_68066328 | 0.16 |
ENSMUST00000063775.5
|
Ldlrad4
|
low density lipoprotein receptor class A domain containing 4 |
| chr2_+_118756973 | 0.16 |
ENSMUST00000099546.6
ENSMUST00000110837.2 |
Chst14
|
carbohydrate sulfotransferase 14 |
| chr2_+_174169492 | 0.15 |
ENSMUST00000156623.8
ENSMUST00000149016.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr8_+_12923771 | 0.15 |
ENSMUST00000156560.2
ENSMUST00000095456.10 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr3_-_51184895 | 0.15 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr17_+_27136065 | 0.15 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr17_-_57023788 | 0.15 |
ENSMUST00000067931.7
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr1_-_39616445 | 0.14 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr1_-_165021879 | 0.14 |
ENSMUST00000043338.10
|
Sft2d2
|
SFT2 domain containing 2 |
| chr1_-_127605660 | 0.14 |
ENSMUST00000160616.8
|
Tmem163
|
transmembrane protein 163 |
| chr7_-_126483851 | 0.14 |
ENSMUST00000071268.11
ENSMUST00000117394.2 |
Taok2
|
TAO kinase 2 |
| chr15_+_78294154 | 0.14 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chrX_-_99670174 | 0.14 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr11_+_118913788 | 0.13 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr2_+_25162487 | 0.13 |
ENSMUST00000028341.11
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr1_-_9770434 | 0.13 |
ENSMUST00000088658.11
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr15_-_74581384 | 0.13 |
ENSMUST00000050234.4
|
Jrk
|
jerky |
| chr2_+_3705824 | 0.12 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr9_-_4309426 | 0.12 |
ENSMUST00000051589.9
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr3_-_90373165 | 0.12 |
ENSMUST00000029540.13
|
Npr1
|
natriuretic peptide receptor 1 |
| chr10_+_108167973 | 0.12 |
ENSMUST00000095313.5
|
Pawr
|
PRKC, apoptosis, WT1, regulator |
| chr6_-_124710084 | 0.12 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_+_108637816 | 0.12 |
ENSMUST00000163617.2
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr8_+_84627332 | 0.12 |
ENSMUST00000045393.15
ENSMUST00000132500.8 ENSMUST00000152978.8 |
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr5_+_114706077 | 0.12 |
ENSMUST00000043650.8
|
Fam222a
|
family with sequence similarity 222, member A |
| chr18_+_38088179 | 0.12 |
ENSMUST00000176902.8
ENSMUST00000176104.8 |
Rell2
|
RELT-like 2 |
| chr9_-_21223631 | 0.12 |
ENSMUST00000115433.11
|
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr11_-_101316156 | 0.11 |
ENSMUST00000103102.10
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr9_+_98868418 | 0.11 |
ENSMUST00000035038.8
ENSMUST00000112911.9 |
Faim
|
Fas apoptotic inhibitory molecule |
| chr1_+_138891447 | 0.11 |
ENSMUST00000168527.8
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr14_+_101891416 | 0.11 |
ENSMUST00000002289.8
|
Uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr5_-_144160397 | 0.11 |
ENSMUST00000085701.7
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr12_-_108145469 | 0.11 |
ENSMUST00000125916.3
ENSMUST00000109879.8 |
Setd3
|
SET domain containing 3 |
| chr4_+_129878890 | 0.11 |
ENSMUST00000106017.8
ENSMUST00000121049.8 |
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr19_-_44058175 | 0.10 |
ENSMUST00000172041.8
ENSMUST00000071698.13 ENSMUST00000112028.10 |
Erlin1
|
ER lipid raft associated 1 |
| chr11_+_44409775 | 0.10 |
ENSMUST00000019333.10
|
Rnf145
|
ring finger protein 145 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hmx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.5 | 1.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.4 | 1.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.7 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 0.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 1.0 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.2 | 3.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 1.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.9 | GO:1905034 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.2 | 1.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.2 | 0.8 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 0.7 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.2 | 0.5 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.1 | 1.4 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 1.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.5 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.5 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.7 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.4 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.3 | GO:0032379 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 0.6 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.7 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.7 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.5 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.8 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.4 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.9 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0032324 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0009092 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.3 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0051466 | positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.2 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.5 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 1.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 0.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.4 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.3 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.0 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.5 | 1.9 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.4 | 2.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 0.5 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 0.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.9 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.1 | 0.5 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 0.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 4.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |