Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hnf1b
Z-value: 0.91
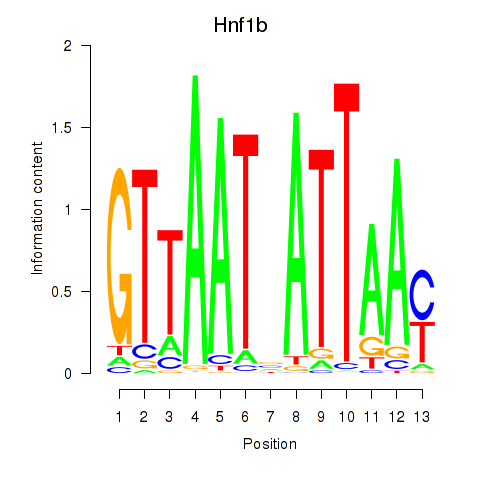
Transcription factors associated with Hnf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1b
|
ENSMUSG00000020679.12 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1b | mm39_v1_chr11_+_83741657_83741681 | 0.71 | 1.8e-01 | Click! |
Activity profile of Hnf1b motif
Sorted Z-values of Hnf1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_87610895 | 1.50 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr2_-_89855921 | 1.06 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264 |
| chr2_+_87609827 | 1.05 |
ENSMUST00000105210.3
|
Olfr152
|
olfactory receptor 152 |
| chr8_-_5155347 | 1.05 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr3_-_66204228 | 0.85 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr6_-_116693849 | 0.74 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr13_-_93810808 | 0.68 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr7_-_85895409 | 0.65 |
ENSMUST00000165771.2
ENSMUST00000233075.2 ENSMUST00000233317.2 ENSMUST00000233312.2 ENSMUST00000233928.2 ENSMUST00000232799.2 ENSMUST00000233744.2 |
Vmn2r76
|
vomeronasal 2, receptor 76 |
| chr2_-_34990689 | 0.64 |
ENSMUST00000226631.2
ENSMUST00000045776.5 ENSMUST00000226972.2 |
AI182371
|
expressed sequence AI182371 |
| chr16_+_24266829 | 0.62 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr12_+_76580386 | 0.62 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr17_+_20634408 | 0.61 |
ENSMUST00000233980.2
ENSMUST00000233642.2 ENSMUST00000233743.2 ENSMUST00000233318.2 ENSMUST00000233161.2 ENSMUST00000233891.2 ENSMUST00000233600.2 ENSMUST00000233863.2 |
Vmn1r224
|
vomeronasal 1 receptor 224 |
| chr10_-_89457115 | 0.56 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr19_-_30526916 | 0.56 |
ENSMUST00000025803.9
|
Dkk1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr2_-_111965322 | 0.54 |
ENSMUST00000213696.2
|
Olfr1316
|
olfactory receptor 1316 |
| chr10_-_75946790 | 0.53 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr8_+_117822593 | 0.53 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr7_-_103734672 | 0.51 |
ENSMUST00000057104.7
|
Olfr645
|
olfactory receptor 645 |
| chr7_-_5152669 | 0.50 |
ENSMUST00000236378.2
|
Vmn1r55
|
vomeronasal 1 receptor 55 |
| chr17_-_37974666 | 0.49 |
ENSMUST00000215414.2
ENSMUST00000213638.3 |
Olfr117
|
olfactory receptor 117 |
| chr7_+_119773070 | 0.46 |
ENSMUST00000033201.7
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr9_-_19088608 | 0.46 |
ENSMUST00000212127.3
|
Olfr839-ps1
|
olfactory receptor 839, pseudogene 1 |
| chr2_-_111400026 | 0.43 |
ENSMUST00000217772.2
ENSMUST00000207283.3 |
Olfr1295
|
olfactory receptor 1295 |
| chr19_+_13208692 | 0.42 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463 |
| chr12_-_103597663 | 0.42 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr4_-_139695337 | 0.41 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr1_+_40478787 | 0.39 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr7_+_23274951 | 0.37 |
ENSMUST00000228832.2
ENSMUST00000227547.2 ENSMUST00000226669.2 ENSMUST00000227932.2 |
Vmn1r169
|
vomeronasal 1 receptor 169 |
| chr11_-_73742280 | 0.36 |
ENSMUST00000213365.2
|
Olfr393
|
olfactory receptor 393 |
| chr8_+_71176713 | 0.34 |
ENSMUST00000034307.14
ENSMUST00000239435.2 ENSMUST00000239487.2 ENSMUST00000110095.3 |
Pde4c
|
phosphodiesterase 4C, cAMP specific |
| chr17_+_18269686 | 0.34 |
ENSMUST00000176802.3
|
Vmn2r124
|
vomeronasal 2, receptor 124 |
| chr9_+_119978773 | 0.33 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr4_+_19818718 | 0.32 |
ENSMUST00000035890.8
|
Slc7a13
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 |
| chr2_-_88581690 | 0.32 |
ENSMUST00000215179.3
ENSMUST00000215529.3 |
Olfr1198
|
olfactory receptor 1198 |
| chr10_-_7162196 | 0.31 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr6_+_56947258 | 0.31 |
ENSMUST00000226130.2
ENSMUST00000228276.2 |
Vmn1r5
|
vomeronasal 1 receptor 5 |
| chr5_+_114427227 | 0.31 |
ENSMUST00000169347.6
|
Myo1h
|
myosin 1H |
| chr11_+_115802828 | 0.29 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr16_+_43067641 | 0.29 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_-_104991477 | 0.28 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr16_+_57173456 | 0.28 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_+_48624295 | 0.27 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr2_-_87838612 | 0.27 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr6_-_137548004 | 0.27 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr11_-_97913420 | 0.26 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr6_-_41681273 | 0.25 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr7_-_139982831 | 0.25 |
ENSMUST00000080153.4
ENSMUST00000216053.2 ENSMUST00000217167.2 |
Olfr531
|
olfactory receptor 531 |
| chr1_+_130659700 | 0.25 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr19_+_42034231 | 0.24 |
ENSMUST00000172244.8
ENSMUST00000081714.5 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr5_-_109372574 | 0.24 |
ENSMUST00000170341.3
|
Vmn2r14
|
vomeronasal 2, receptor 14 |
| chr7_-_29204812 | 0.23 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chrM_+_2743 | 0.22 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr15_-_50753792 | 0.22 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr7_-_102507962 | 0.22 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr4_-_41045381 | 0.22 |
ENSMUST00000054945.8
|
Aqp7
|
aquaporin 7 |
| chr2_+_89821565 | 0.21 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr11_+_110858842 | 0.20 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_+_48279794 | 0.19 |
ENSMUST00000030029.10
|
Invs
|
inversin |
| chr17_+_48047955 | 0.19 |
ENSMUST00000086932.10
|
Tfeb
|
transcription factor EB |
| chr10_+_62897353 | 0.19 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr2_-_86280934 | 0.18 |
ENSMUST00000216162.3
ENSMUST00000217586.3 ENSMUST00000213789.3 |
Olfr1065
|
olfactory receptor 1065 |
| chr3_-_133250889 | 0.18 |
ENSMUST00000197118.5
|
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr4_-_52859227 | 0.18 |
ENSMUST00000107670.3
|
Olfr273
|
olfactory receptor 273 |
| chr5_-_108823435 | 0.17 |
ENSMUST00000051757.14
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr19_-_14575395 | 0.16 |
ENSMUST00000052011.15
ENSMUST00000167776.3 |
Tle4
|
transducin-like enhancer of split 4 |
| chr9_+_99511549 | 0.16 |
ENSMUST00000131095.8
ENSMUST00000078367.12 ENSMUST00000112885.9 |
Dzip1l
|
DAZ interacting protein 1-like |
| chr10_-_95678786 | 0.16 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr12_+_59142439 | 0.16 |
ENSMUST00000219140.3
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr3_-_144525255 | 0.16 |
ENSMUST00000029929.12
|
Clca3a2
|
chloride channel accessory 3A2 |
| chr2_-_88157559 | 0.16 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175 |
| chr9_+_123596276 | 0.15 |
ENSMUST00000166236.9
ENSMUST00000111454.4 ENSMUST00000168910.2 |
Ccr9
|
chemokine (C-C motif) receptor 9 |
| chr4_-_119217079 | 0.14 |
ENSMUST00000143494.3
ENSMUST00000154606.9 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr17_+_37710293 | 0.14 |
ENSMUST00000216844.2
ENSMUST00000215974.2 ENSMUST00000215894.2 |
Olfr107
|
olfactory receptor 107 |
| chr10_-_8761777 | 0.14 |
ENSMUST00000015449.6
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr3_-_116506345 | 0.14 |
ENSMUST00000169530.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr10_-_130230201 | 0.13 |
ENSMUST00000094502.6
ENSMUST00000233146.2 |
Vmn2r84
|
vomeronasal 2, receptor 84 |
| chr5_-_100521343 | 0.13 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr13_-_112717196 | 0.13 |
ENSMUST00000051756.8
|
Il31ra
|
interleukin 31 receptor A |
| chr2_+_162829422 | 0.13 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr8_-_105350898 | 0.13 |
ENSMUST00000212882.2
ENSMUST00000163783.4 |
Cdh16
|
cadherin 16 |
| chr7_-_108484213 | 0.13 |
ENSMUST00000217803.2
|
Olfr518
|
olfactory receptor 518 |
| chr9_+_67747668 | 0.12 |
ENSMUST00000077879.7
|
Vps13c
|
vacuolar protein sorting 13C |
| chr2_+_87854404 | 0.12 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr7_-_44711771 | 0.11 |
ENSMUST00000210101.2
ENSMUST00000209219.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr17_-_33937565 | 0.11 |
ENSMUST00000174040.2
ENSMUST00000173015.8 ENSMUST00000066121.13 ENSMUST00000186022.7 ENSMUST00000173329.8 ENSMUST00000172767.9 |
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chr11_+_46462363 | 0.11 |
ENSMUST00000050937.7
|
BC053393
|
cDNA sequence BC053393 |
| chr6_-_102441628 | 0.11 |
ENSMUST00000032159.7
|
Cntn3
|
contactin 3 |
| chr11_+_116739727 | 0.11 |
ENSMUST00000143184.2
|
Mettl23
|
methyltransferase like 23 |
| chr2_+_3771709 | 0.11 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr18_+_4993795 | 0.11 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr14_-_20546848 | 0.11 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr5_+_107645626 | 0.10 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr9_+_39848797 | 0.10 |
ENSMUST00000213246.3
|
Olfr974
|
olfactory receptor 974 |
| chr15_+_99291491 | 0.10 |
ENSMUST00000159531.3
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr14_-_36641270 | 0.10 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_171954316 | 0.10 |
ENSMUST00000075895.9
ENSMUST00000111252.4 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr10_-_95678748 | 0.10 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr19_-_10460238 | 0.09 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr18_+_47054867 | 0.09 |
ENSMUST00000234910.2
|
Arl14epl
|
ADP-ribosylation factor-like 14 effector protein-like |
| chr19_-_8382424 | 0.09 |
ENSMUST00000064507.12
ENSMUST00000120540.2 ENSMUST00000096269.11 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr3_-_116506294 | 0.09 |
ENSMUST00000029569.9
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr1_+_58834532 | 0.09 |
ENSMUST00000027189.15
|
Casp8
|
caspase 8 |
| chr17_-_35351026 | 0.08 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr7_-_5808444 | 0.08 |
ENSMUST00000075085.7
|
Vmn1r63
|
vomeronasal 1 receptor 63 |
| chr7_-_119494669 | 0.08 |
ENSMUST00000098080.9
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr11_-_54140462 | 0.08 |
ENSMUST00000019060.6
|
Csf2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr10_-_125225298 | 0.08 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr9_-_106035332 | 0.07 |
ENSMUST00000112543.9
|
Glyctk
|
glycerate kinase |
| chr2_-_178049375 | 0.07 |
ENSMUST00000081134.10
|
Sycp2
|
synaptonemal complex protein 2 |
| chr5_-_123804745 | 0.07 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr2_-_87881473 | 0.07 |
ENSMUST00000183862.3
|
Olfr1162
|
olfactory receptor 1162 |
| chr9_+_99511876 | 0.07 |
ENSMUST00000112886.9
|
Dzip1l
|
DAZ interacting protein 1-like |
| chr13_-_73826124 | 0.06 |
ENSMUST00000022105.15
ENSMUST00000109680.10 ENSMUST00000221026.2 ENSMUST00000109679.4 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr10_+_43455919 | 0.06 |
ENSMUST00000214476.2
|
Cd24a
|
CD24a antigen |
| chr7_-_44753168 | 0.06 |
ENSMUST00000211085.2
ENSMUST00000210642.2 ENSMUST00000003512.9 |
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr16_-_19241884 | 0.06 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chrX_-_161747552 | 0.06 |
ENSMUST00000038769.3
|
S100g
|
S100 calcium binding protein G |
| chr17_+_37710117 | 0.05 |
ENSMUST00000215947.2
|
Olfr107
|
olfactory receptor 107 |
| chr1_+_19279138 | 0.05 |
ENSMUST00000027059.11
|
Tfap2b
|
transcription factor AP-2 beta |
| chr1_+_178233640 | 0.05 |
ENSMUST00000027775.9
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr11_-_75330302 | 0.05 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr11_+_58643560 | 0.05 |
ENSMUST00000215071.2
|
Olfr316
|
olfactory receptor 316 |
| chr9_-_48876290 | 0.05 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr13_+_23991010 | 0.05 |
ENSMUST00000006786.11
ENSMUST00000099697.3 |
Slc17a2
|
solute carrier family 17 (sodium phosphate), member 2 |
| chr15_-_79169671 | 0.04 |
ENSMUST00000170955.2
ENSMUST00000165408.8 |
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr12_+_108572015 | 0.04 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr11_-_75330415 | 0.04 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr4_+_119494901 | 0.03 |
ENSMUST00000024015.3
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr14_-_79718890 | 0.03 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr14_+_47120311 | 0.03 |
ENSMUST00000022386.15
ENSMUST00000228404.2 ENSMUST00000100672.11 |
Samd4
|
sterile alpha motif domain containing 4 |
| chr19_+_43770619 | 0.03 |
ENSMUST00000026208.6
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr14_-_36641470 | 0.03 |
ENSMUST00000182042.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr19_+_5928649 | 0.03 |
ENSMUST00000136833.8
ENSMUST00000141362.2 |
Slc25a45
|
solute carrier family 25, member 45 |
| chrX_+_100492684 | 0.03 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr14_+_54491637 | 0.03 |
ENSMUST00000180359.8
ENSMUST00000199338.2 |
Abhd4
|
abhydrolase domain containing 4 |
| chr9_-_106035308 | 0.03 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr7_-_109271433 | 0.02 |
ENSMUST00000207394.2
|
Denn2b
|
DENN domain containing 2B |
| chr9_+_72866067 | 0.02 |
ENSMUST00000098567.9
ENSMUST00000034734.9 |
Dnaaf4
|
dynein axonemal assembly factor 4 |
| chr17_+_74702601 | 0.02 |
ENSMUST00000024870.9
ENSMUST00000179074.9 ENSMUST00000233799.2 ENSMUST00000233042.2 |
Slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr6_+_142291374 | 0.02 |
ENSMUST00000041852.8
|
Pyroxd1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr9_+_21746785 | 0.02 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr1_+_133237516 | 0.02 |
ENSMUST00000094557.7
ENSMUST00000192465.2 ENSMUST00000193888.6 ENSMUST00000194044.6 ENSMUST00000184603.8 |
Golt1a
Gm28040
Gm28040
|
golgi transport 1A predicted gene, 28040 predicted gene, 28040 |
| chr1_-_36312482 | 0.02 |
ENSMUST00000056946.8
|
Neurl3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr12_-_76842263 | 0.02 |
ENSMUST00000082431.6
|
Gpx2
|
glutathione peroxidase 2 |
| chr4_+_43957401 | 0.02 |
ENSMUST00000030202.14
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr14_+_61547202 | 0.01 |
ENSMUST00000055159.8
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr6_+_70549568 | 0.01 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr11_-_6180127 | 0.01 |
ENSMUST00000004505.3
|
Npc1l1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr6_-_129853617 | 0.01 |
ENSMUST00000014687.11
ENSMUST00000122219.2 |
Klra17
|
killer cell lectin-like receptor, subfamily A, member 17 |
| chr6_+_38895902 | 0.01 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr17_-_35265514 | 0.01 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr14_+_26616514 | 0.01 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr17_+_23879448 | 0.01 |
ENSMUST00000062967.10
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr13_-_53440087 | 0.01 |
ENSMUST00000021918.10
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr7_+_102731092 | 0.01 |
ENSMUST00000214215.2
|
Olfr584
|
olfactory receptor 584 |
| chr13_+_55300453 | 0.01 |
ENSMUST00000005452.6
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr4_-_6275629 | 0.01 |
ENSMUST00000029905.2
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr4_-_140344373 | 0.01 |
ENSMUST00000154979.2
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr5_-_107074110 | 0.01 |
ENSMUST00000117588.8
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr13_+_54849268 | 0.01 |
ENSMUST00000037145.8
|
Cdhr2
|
cadherin-related family member 2 |
| chr19_+_53781721 | 0.01 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr13_+_93810911 | 0.00 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr3_-_14676309 | 0.00 |
ENSMUST00000185423.2
ENSMUST00000186870.7 ENSMUST00000185384.7 |
Rbis
|
ribosomal biogenesis factor |
| chr16_-_20972750 | 0.00 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr7_-_83444026 | 0.00 |
ENSMUST00000119134.8
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr9_+_54446268 | 0.00 |
ENSMUST00000060242.12
|
Sh2d7
|
SH2 domain containing 7 |
| chr1_-_172722589 | 0.00 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr6_-_38101503 | 0.00 |
ENSMUST00000040259.8
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr14_-_36832044 | 0.00 |
ENSMUST00000179488.3
|
2610528A11Rik
|
RIKEN cDNA 2610528A11 gene |
| chr1_+_125604706 | 0.00 |
ENSMUST00000027581.7
|
Gpr39
|
G protein-coupled receptor 39 |
| chr3_-_108133914 | 0.00 |
ENSMUST00000141387.4
|
Sypl2
|
synaptophysin-like 2 |
| chr17_+_57369231 | 0.00 |
ENSMUST00000097299.10
ENSMUST00000169543.8 ENSMUST00000163763.2 |
Crb3
|
crumbs family member 3 |
| chr14_+_33807964 | 0.00 |
ENSMUST00000120077.2
|
Anxa8
|
annexin A8 |
| chr7_-_107400415 | 0.00 |
ENSMUST00000106755.3
|
Ovch2
|
ovochymase 2 |
| chr11_-_75828504 | 0.00 |
ENSMUST00000108420.9
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr8_+_123920682 | 0.00 |
ENSMUST00000212409.2
|
Dpep1
|
dipeptidase 1 |
| chr7_-_83444050 | 0.00 |
ENSMUST00000011298.14
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr14_+_33807935 | 0.00 |
ENSMUST00000022519.15
|
Anxa8
|
annexin A8 |
| chr3_+_69129745 | 0.00 |
ENSMUST00000183126.2
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr1_-_92419475 | 0.00 |
ENSMUST00000071521.3
|
Olfr1415
|
olfactory receptor 1415 |
| chr11_+_70410009 | 0.00 |
ENSMUST00000057685.3
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904956 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.7 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.5 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.2 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.2 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.2 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 | 0.2 | GO:0015793 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.0 | 0.1 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0051325 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.0 | 0.1 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.0 | GO:0097275 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |