Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
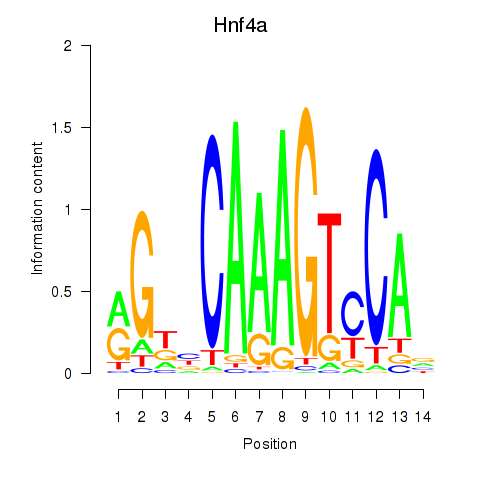
Results for Hnf4a
Z-value: 0.83
Transcription factors associated with Hnf4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf4a
|
ENSMUSG00000017950.17 | hepatic nuclear factor 4, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf4a | mm39_v1_chr2_+_163348728_163348780 | 0.86 | 6.5e-02 | Click! |
Activity profile of Hnf4a motif
Sorted Z-values of Hnf4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_135339543 | 0.68 |
ENSMUST00000205156.3
|
Emp1
|
epithelial membrane protein 1 |
| chr12_+_113115632 | 0.46 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr7_-_19410749 | 0.41 |
ENSMUST00000003074.16
|
Apoc2
|
apolipoprotein C-II |
| chr14_-_55995912 | 0.40 |
ENSMUST00000001497.9
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr4_-_138053603 | 0.36 |
ENSMUST00000030536.13
|
Pink1
|
PTEN induced putative kinase 1 |
| chr10_-_127016448 | 0.35 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr17_-_71297885 | 0.33 |
ENSMUST00000038446.10
|
Myl12b
|
myosin, light chain 12B, regulatory |
| chr11_+_115768323 | 0.32 |
ENSMUST00000222123.2
|
Myo15b
|
myosin XVB |
| chr15_-_76501525 | 0.27 |
ENSMUST00000230977.2
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr11_-_69304501 | 0.27 |
ENSMUST00000094077.5
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr1_+_131936022 | 0.25 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr4_-_138053545 | 0.25 |
ENSMUST00000105817.4
|
Pink1
|
PTEN induced putative kinase 1 |
| chr8_-_13250535 | 0.24 |
ENSMUST00000165605.4
ENSMUST00000209691.2 ENSMUST00000211128.2 ENSMUST00000210317.2 |
Grtp1
|
GH regulated TBC protein 1 |
| chr4_+_134658209 | 0.23 |
ENSMUST00000030622.3
|
Syf2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae) |
| chr15_+_3300249 | 0.23 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr4_-_132149704 | 0.23 |
ENSMUST00000152271.8
ENSMUST00000084170.12 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr5_-_137869969 | 0.23 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr7_+_29931309 | 0.21 |
ENSMUST00000019882.16
ENSMUST00000149654.8 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr1_+_157353696 | 0.21 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr5_+_31855304 | 0.21 |
ENSMUST00000114515.9
|
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr1_+_74324089 | 0.21 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr7_+_27151838 | 0.20 |
ENSMUST00000108357.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr12_+_59178258 | 0.20 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr8_-_84321032 | 0.19 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr12_+_59178072 | 0.18 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr15_-_102630589 | 0.18 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr8_-_84321069 | 0.18 |
ENSMUST00000019382.17
ENSMUST00000212630.2 ENSMUST00000165740.9 |
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr5_+_31855009 | 0.17 |
ENSMUST00000201352.4
ENSMUST00000202815.4 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr11_-_4654303 | 0.17 |
ENSMUST00000058407.6
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr16_+_20492564 | 0.16 |
ENSMUST00000141034.8
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr13_-_73476561 | 0.16 |
ENSMUST00000222930.2
ENSMUST00000223293.2 ENSMUST00000022097.6 |
Ndufs6
|
NADH:ubiquinone oxidoreductase core subunit S6 |
| chr15_-_103242697 | 0.16 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr9_+_21914083 | 0.15 |
ENSMUST00000216344.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_95041246 | 0.14 |
ENSMUST00000172572.9
ENSMUST00000173462.3 |
Scnm1
|
sodium channel modifier 1 |
| chr7_-_19684654 | 0.14 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chrX_+_35592006 | 0.14 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr1_-_133352115 | 0.14 |
ENSMUST00000153799.8
|
Sox13
|
SRY (sex determining region Y)-box 13 |
| chr10_+_81395242 | 0.14 |
ENSMUST00000002518.9
|
Tle5
|
TLE family member 5, transcriptional modulator |
| chr9_+_21914296 | 0.13 |
ENSMUST00000003493.9
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr8_+_3637785 | 0.13 |
ENSMUST00000171962.3
ENSMUST00000207712.2 ENSMUST00000207970.2 ENSMUST00000207533.2 ENSMUST00000208240.2 ENSMUST00000207432.2 ENSMUST00000207077.2 |
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr6_-_86742789 | 0.13 |
ENSMUST00000123732.4
|
Anxa4
|
annexin A4 |
| chr6_-_86742847 | 0.12 |
ENSMUST00000113675.8
|
Anxa4
|
annexin A4 |
| chr17_-_74257164 | 0.12 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr16_+_20492861 | 0.12 |
ENSMUST00000151679.3
|
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr5_+_31855394 | 0.12 |
ENSMUST00000063813.11
ENSMUST00000071531.12 ENSMUST00000131995.7 ENSMUST00000114507.10 |
Babam2
|
BRISC and BRCA1 A complex member 2 |
| chr2_+_90927053 | 0.12 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr9_-_110818679 | 0.12 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr5_-_104125226 | 0.11 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr7_+_29931735 | 0.11 |
ENSMUST00000108193.2
ENSMUST00000108192.2 |
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr2_-_13496624 | 0.11 |
ENSMUST00000091436.7
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr14_-_70414236 | 0.11 |
ENSMUST00000153735.8
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr17_-_36220518 | 0.11 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr3_+_107784543 | 0.11 |
ENSMUST00000037375.10
ENSMUST00000199990.2 |
Eps8l3
|
EPS8-like 3 |
| chr7_-_100661181 | 0.11 |
ENSMUST00000178340.3
ENSMUST00000037540.5 |
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr12_+_59177552 | 0.11 |
ENSMUST00000175877.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr8_+_12965876 | 0.11 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr5_-_24652775 | 0.11 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr9_+_21914334 | 0.11 |
ENSMUST00000115331.10
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr15_-_78739717 | 0.10 |
ENSMUST00000044584.6
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr1_+_87983099 | 0.10 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr17_+_34824827 | 0.10 |
ENSMUST00000037489.15
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr9_+_62765362 | 0.09 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr10_-_13428855 | 0.09 |
ENSMUST00000105539.2
|
Pex3
|
peroxisomal biogenesis factor 3 |
| chr2_-_77776524 | 0.09 |
ENSMUST00000111824.8
ENSMUST00000111819.8 ENSMUST00000128963.2 |
Cwc22
|
CWC22 spliceosome-associated protein |
| chr4_+_152284261 | 0.09 |
ENSMUST00000105652.3
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr15_+_78294154 | 0.09 |
ENSMUST00000229739.2
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr4_+_53631460 | 0.09 |
ENSMUST00000132151.8
ENSMUST00000159415.9 ENSMUST00000163067.9 |
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr16_-_16377982 | 0.09 |
ENSMUST00000161861.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr7_-_45408951 | 0.08 |
ENSMUST00000075571.16
|
Sult2b1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chr7_-_110213968 | 0.08 |
ENSMUST00000166020.8
ENSMUST00000171218.8 ENSMUST00000033058.14 ENSMUST00000164759.8 |
Sbf2
|
SET binding factor 2 |
| chr2_+_157870399 | 0.08 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr10_-_20600442 | 0.08 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr15_-_89064936 | 0.08 |
ENSMUST00000109331.9
|
Plxnb2
|
plexin B2 |
| chr7_-_100661220 | 0.08 |
ENSMUST00000207916.2
|
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr6_-_122579320 | 0.08 |
ENSMUST00000147760.8
ENSMUST00000112585.8 ENSMUST00000203309.3 |
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr19_+_53186430 | 0.08 |
ENSMUST00000237099.2
|
Add3
|
adducin 3 (gamma) |
| chr12_-_80159768 | 0.08 |
ENSMUST00000219642.2
ENSMUST00000165114.2 ENSMUST00000218835.2 ENSMUST00000021552.3 |
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr4_-_148529187 | 0.08 |
ENSMUST00000051633.3
|
Ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr3_+_107186309 | 0.08 |
ENSMUST00000199317.2
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr4_+_118266526 | 0.07 |
ENSMUST00000084319.11
ENSMUST00000106384.10 ENSMUST00000126089.8 ENSMUST00000073881.8 ENSMUST00000019229.15 |
Med8
|
mediator complex subunit 8 |
| chr9_+_21914513 | 0.07 |
ENSMUST00000215795.2
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr19_+_4771089 | 0.07 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr11_-_3454766 | 0.07 |
ENSMUST00000044507.12
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr17_-_34822649 | 0.07 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr19_+_10019023 | 0.07 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr8_+_117822593 | 0.07 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr8_-_122671588 | 0.07 |
ENSMUST00000057653.8
|
Car5a
|
carbonic anhydrase 5a, mitochondrial |
| chr16_-_45474413 | 0.07 |
ENSMUST00000036732.9
|
BC016579
|
cDNA sequence, BC016579 |
| chr19_+_5138562 | 0.06 |
ENSMUST00000238093.2
ENSMUST00000025811.6 ENSMUST00000237025.2 |
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr2_+_112069813 | 0.06 |
ENSMUST00000028554.4
|
Lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr9_-_66951114 | 0.06 |
ENSMUST00000113686.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr5_-_137856280 | 0.06 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr15_+_12117899 | 0.06 |
ENSMUST00000122941.8
|
Zfr
|
zinc finger RNA binding protein |
| chr5_+_20112500 | 0.06 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_-_37178079 | 0.06 |
ENSMUST00000025329.13
ENSMUST00000174195.8 |
Trim15
|
tripartite motif-containing 15 |
| chr10_+_94386714 | 0.06 |
ENSMUST00000148910.3
ENSMUST00000117460.8 |
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr19_-_45986919 | 0.06 |
ENSMUST00000045396.9
|
Armh3
|
armadillo-like helical domain containing 3 |
| chr6_-_122579362 | 0.06 |
ENSMUST00000112586.8
|
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr5_+_114284585 | 0.05 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr9_+_58489523 | 0.05 |
ENSMUST00000177292.8
ENSMUST00000085651.12 ENSMUST00000176557.8 ENSMUST00000114121.11 ENSMUST00000177064.8 |
Nptn
|
neuroplastin |
| chr12_+_72583114 | 0.05 |
ENSMUST00000044352.8
|
Pcnx4
|
pecanex homolog 4 |
| chr19_+_8828132 | 0.05 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr12_+_21366386 | 0.05 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr5_-_137834470 | 0.05 |
ENSMUST00000110980.2
ENSMUST00000058897.11 ENSMUST00000199028.2 |
Pilra
|
paired immunoglobin-like type 2 receptor alpha |
| chr2_+_70948267 | 0.04 |
ENSMUST00000028403.3
|
Cybrd1
|
cytochrome b reductase 1 |
| chr7_+_143027473 | 0.04 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr14_-_24054927 | 0.04 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr15_-_82648376 | 0.04 |
ENSMUST00000055721.6
|
Cyp2d40
|
cytochrome P450, family 2, subfamily d, polypeptide 40 |
| chr7_+_30758767 | 0.04 |
ENSMUST00000039775.9
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr7_-_135130374 | 0.04 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr2_+_25390762 | 0.04 |
ENSMUST00000015239.10
|
Fbxw5
|
F-box and WD-40 domain protein 5 |
| chr9_-_66950991 | 0.04 |
ENSMUST00000113689.8
ENSMUST00000113684.8 |
Tpm1
|
tropomyosin 1, alpha |
| chr17_-_26240827 | 0.04 |
ENSMUST00000118828.8
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr3_+_107186151 | 0.04 |
ENSMUST00000145735.6
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr8_-_41087793 | 0.04 |
ENSMUST00000173957.2
ENSMUST00000048898.17 ENSMUST00000174205.8 |
Mtmr7
|
myotubularin related protein 7 |
| chr1_-_155293075 | 0.04 |
ENSMUST00000027741.12
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr5_-_137870001 | 0.04 |
ENSMUST00000164886.2
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr7_-_83384711 | 0.04 |
ENSMUST00000001792.12
|
Il16
|
interleukin 16 |
| chr8_-_95929420 | 0.04 |
ENSMUST00000212842.2
|
Kifc3
|
kinesin family member C3 |
| chrX_-_72703330 | 0.04 |
ENSMUST00000114473.8
ENSMUST00000002087.14 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr2_-_58990967 | 0.04 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr11_+_73090270 | 0.04 |
ENSMUST00000006105.7
|
Shpk
|
sedoheptulokinase |
| chr2_-_155424576 | 0.04 |
ENSMUST00000126322.8
|
Gss
|
glutathione synthetase |
| chr4_-_123507494 | 0.04 |
ENSMUST00000238866.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_+_110427643 | 0.03 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr15_-_76004395 | 0.03 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr1_-_136161850 | 0.03 |
ENSMUST00000120339.8
|
Inava
|
innate immunity activator |
| chr8_+_105267431 | 0.03 |
ENSMUST00000056051.11
|
Car7
|
carbonic anhydrase 7 |
| chr2_-_77776719 | 0.03 |
ENSMUST00000065889.10
|
Cwc22
|
CWC22 spliceosome-associated protein |
| chr15_-_90934021 | 0.03 |
ENSMUST00000109287.4
ENSMUST00000067205.16 ENSMUST00000088614.13 |
Kif21a
|
kinesin family member 21A |
| chr9_-_66951025 | 0.03 |
ENSMUST00000113695.8
|
Tpm1
|
tropomyosin 1, alpha |
| chr1_-_74076279 | 0.03 |
ENSMUST00000187281.7
|
Tns1
|
tensin 1 |
| chr19_+_46140942 | 0.03 |
ENSMUST00000026254.14
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr12_-_115157739 | 0.03 |
ENSMUST00000103525.3
|
Ighv1-54
|
immunoglobulin heavy variable V1-54 |
| chr12_+_37291632 | 0.03 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr17_-_37794434 | 0.03 |
ENSMUST00000016427.11
ENSMUST00000171139.3 |
H2-M2
|
histocompatibility 2, M region locus 2 |
| chr11_+_78389913 | 0.03 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr19_-_11058452 | 0.03 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr7_-_28092113 | 0.03 |
ENSMUST00000003536.9
|
Med29
|
mediator complex subunit 29 |
| chr1_+_185187000 | 0.03 |
ENSMUST00000061093.7
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr12_+_31123860 | 0.03 |
ENSMUST00000041133.10
|
Fam110c
|
family with sequence similarity 110, member C |
| chr11_-_86648309 | 0.03 |
ENSMUST00000060766.16
ENSMUST00000103186.11 |
Cltc
|
clathrin, heavy polypeptide (Hc) |
| chr16_-_30086317 | 0.03 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr11_+_22462088 | 0.03 |
ENSMUST00000059319.8
|
Tmem17
|
transmembrane protein 17 |
| chr4_+_95467653 | 0.03 |
ENSMUST00000043335.11
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr4_-_41731142 | 0.03 |
ENSMUST00000171251.8
|
Arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr4_+_141473983 | 0.03 |
ENSMUST00000038161.5
|
Agmat
|
agmatine ureohydrolase (agmatinase) |
| chr7_+_28092370 | 0.03 |
ENSMUST00000003529.9
|
Paf1
|
Paf1, RNA polymerase II complex component |
| chr9_-_21913833 | 0.03 |
ENSMUST00000115336.10
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr15_-_82505132 | 0.03 |
ENSMUST00000109515.3
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr3_+_95041399 | 0.03 |
ENSMUST00000066386.6
|
Lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr19_-_7272758 | 0.03 |
ENSMUST00000025921.15
|
Mark2
|
MAP/microtubule affinity regulating kinase 2 |
| chr4_-_126057263 | 0.03 |
ENSMUST00000097891.4
|
Sh3d21
|
SH3 domain containing 21 |
| chr12_+_51640097 | 0.03 |
ENSMUST00000164782.10
ENSMUST00000085412.7 |
Coch
|
cochlin |
| chr15_+_82336535 | 0.03 |
ENSMUST00000089129.7
ENSMUST00000229313.2 ENSMUST00000231136.2 |
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9 |
| chr18_+_36481706 | 0.03 |
ENSMUST00000235864.2
ENSMUST00000050584.10 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr7_+_89780785 | 0.03 |
ENSMUST00000208684.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_-_52257452 | 0.03 |
ENSMUST00000228162.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_-_10400710 | 0.03 |
ENSMUST00000078748.4
|
Slc10a5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5 |
| chr4_-_62005498 | 0.03 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_88595161 | 0.02 |
ENSMUST00000105148.2
|
Ifna16
|
interferon alpha 16 |
| chr15_-_90934059 | 0.02 |
ENSMUST00000109288.9
ENSMUST00000100304.11 |
Kif21a
|
kinesin family member 21A |
| chr4_-_152216322 | 0.02 |
ENSMUST00000105653.8
|
Espn
|
espin |
| chr12_+_37292029 | 0.02 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr2_+_172994841 | 0.02 |
ENSMUST00000029017.6
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr2_-_77000936 | 0.02 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr19_-_40371016 | 0.02 |
ENSMUST00000225766.3
|
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_+_192984278 | 0.02 |
ENSMUST00000016315.16
|
Lamb3
|
laminin, beta 3 |
| chr4_-_132030643 | 0.02 |
ENSMUST00000040411.7
|
Rab42
|
RAB42, member RAS oncogene family |
| chr18_-_32170012 | 0.02 |
ENSMUST00000134663.2
|
Myo7b
|
myosin VIIB |
| chr17_-_91396154 | 0.02 |
ENSMUST00000161402.10
ENSMUST00000054059.15 ENSMUST00000072671.14 ENSMUST00000174331.8 |
Nrxn1
|
neurexin I |
| chrX_+_10118544 | 0.02 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr11_+_70396070 | 0.02 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr9_-_21913896 | 0.02 |
ENSMUST00000044926.6
|
Odad3
|
outer dynein arm docking complex subunit 3 |
| chr14_+_54713557 | 0.02 |
ENSMUST00000164766.8
|
Rem2
|
rad and gem related GTP binding protein 2 |
| chr18_+_63841756 | 0.02 |
ENSMUST00000072726.7
ENSMUST00000235648.2 ENSMUST00000236879.2 |
Wdr7
|
WD repeat domain 7 |
| chr6_-_71299184 | 0.02 |
ENSMUST00000173297.2
ENSMUST00000114188.3 |
Smyd1
|
SET and MYND domain containing 1 |
| chr11_+_69945157 | 0.02 |
ENSMUST00000108585.9
ENSMUST00000018699.13 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr16_+_37400500 | 0.02 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr10_+_70040483 | 0.02 |
ENSMUST00000020090.8
|
Mrln
|
myoregulin |
| chr5_+_20112771 | 0.02 |
ENSMUST00000200443.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_30193047 | 0.02 |
ENSMUST00000058280.13
ENSMUST00000133318.8 ENSMUST00000142575.8 ENSMUST00000131040.2 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr4_+_110254907 | 0.02 |
ENSMUST00000097920.9
ENSMUST00000080744.13 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr19_+_44980565 | 0.02 |
ENSMUST00000179305.2
|
Sema4g
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr11_-_75330302 | 0.02 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr17_-_46749370 | 0.02 |
ENSMUST00000087012.7
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr15_+_99870661 | 0.02 |
ENSMUST00000100206.4
|
Larp4
|
La ribonucleoprotein domain family, member 4 |
| chr8_-_114575247 | 0.02 |
ENSMUST00000093113.5
|
Adamts18
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 18 |
| chr5_-_137202790 | 0.02 |
ENSMUST00000041226.11
|
Muc3
|
mucin 3, intestinal |
| chr8_-_93956143 | 0.02 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr6_-_41613322 | 0.02 |
ENSMUST00000031902.7
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr7_+_80764564 | 0.02 |
ENSMUST00000119083.2
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr15_+_10358611 | 0.02 |
ENSMUST00000110541.8
ENSMUST00000110542.8 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr16_-_17745999 | 0.02 |
ENSMUST00000003622.16
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr2_+_3705824 | 0.02 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr7_+_80764547 | 0.02 |
ENSMUST00000026820.11
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr12_+_37291728 | 0.02 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr11_+_50248586 | 0.02 |
ENSMUST00000052596.3
|
Cby3
|
chibby family member 3 |
| chr2_-_77000878 | 0.02 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr1_-_90897329 | 0.02 |
ENSMUST00000130042.2
ENSMUST00000027529.12 |
Rab17
|
RAB17, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.6 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of macromitophagy(GO:1901526) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0090366 | DNA cytosine deamination(GO:0070383) positive regulation of mRNA modification(GO:0090366) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0042761 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.7 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.0 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.2 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.0 | 0.1 | GO:0004131 | cytosine deaminase activity(GO:0004131) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0050294 | alcohol sulfotransferase activity(GO:0004027) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |