Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Homez
Z-value: 2.50
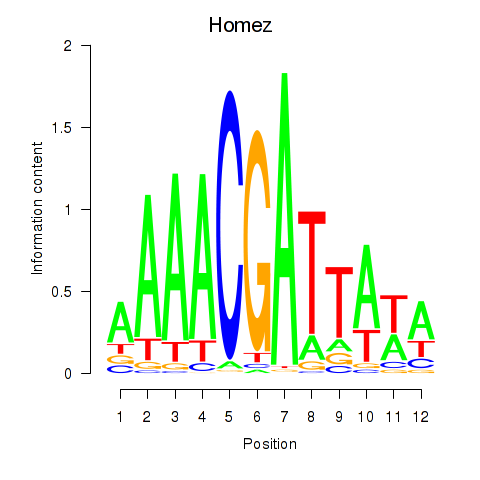
Transcription factors associated with Homez
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Homez
|
ENSMUSG00000057156.11 | homeodomain leucine zipper-encoding gene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Homez | mm39_v1_chr14_-_55108384_55108418 | 0.95 | 1.3e-02 | Click! |
Activity profile of Homez motif
Sorted Z-values of Homez motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_151586063 | 1.77 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr4_-_117035922 | 1.56 |
ENSMUST00000153953.2
ENSMUST00000106436.8 |
Kif2c
|
kinesin family member 2C |
| chr8_-_34578880 | 1.32 |
ENSMUST00000080152.5
|
Gm10131
|
predicted pseudogene 10131 |
| chr15_-_38518406 | 1.21 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr4_+_102617332 | 1.20 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr19_-_6885657 | 1.14 |
ENSMUST00000149261.8
|
Prdx5
|
peroxiredoxin 5 |
| chr11_-_40586029 | 1.08 |
ENSMUST00000101347.10
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr7_+_140462343 | 1.08 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr16_+_49620883 | 1.03 |
ENSMUST00000229640.2
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr17_+_37581103 | 1.01 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr2_-_86180622 | 0.99 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr1_-_118239146 | 0.96 |
ENSMUST00000027623.9
|
Tsn
|
translin |
| chr18_-_42395207 | 0.96 |
ENSMUST00000097590.5
|
Lars
|
leucyl-tRNA synthetase |
| chr7_-_47139334 | 0.93 |
ENSMUST00000119798.3
|
Mrgpra2b
|
MAS-related GPR, member A2B |
| chr7_+_140461860 | 0.93 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr3_-_130503041 | 0.92 |
ENSMUST00000043937.9
|
Ostc
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr10_-_112764879 | 0.90 |
ENSMUST00000099276.4
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr4_-_116851550 | 0.89 |
ENSMUST00000130273.8
|
Urod
|
uroporphyrinogen decarboxylase |
| chr5_-_45607485 | 0.86 |
ENSMUST00000154962.8
ENSMUST00000118097.8 ENSMUST00000198258.5 |
Qdpr
|
quinoid dihydropteridine reductase |
| chr8_-_4829519 | 0.85 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr2_+_71219561 | 0.82 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr8_-_108151661 | 0.82 |
ENSMUST00000003946.9
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog |
| chr3_-_100069680 | 0.82 |
ENSMUST00000052120.14
|
Wdr3
|
WD repeat domain 3 |
| chr14_-_118370144 | 0.81 |
ENSMUST00000022727.10
ENSMUST00000228543.2 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr10_-_115198093 | 0.79 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr14_+_20344765 | 0.78 |
ENSMUST00000223663.2
ENSMUST00000022343.6 ENSMUST00000224066.2 ENSMUST00000223941.2 ENSMUST00000224311.2 |
Nudt13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr3_-_50398027 | 0.78 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr1_-_162376053 | 0.78 |
ENSMUST00000028017.16
|
Eef1aknmt
|
EEF1A lysine and N-terminal methyltransferase |
| chr11_-_48762170 | 0.77 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr2_-_126342551 | 0.76 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr12_-_103392039 | 0.74 |
ENSMUST00000110001.4
ENSMUST00000223233.2 ENSMUST00000044923.15 ENSMUST00000221211.2 |
Ddx24
|
DEAD box helicase 24 |
| chr2_-_23939401 | 0.71 |
ENSMUST00000051416.12
|
Hnmt
|
histamine N-methyltransferase |
| chr18_+_84106188 | 0.70 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr12_-_40298072 | 0.69 |
ENSMUST00000169926.8
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chrX_-_135644424 | 0.68 |
ENSMUST00000166478.8
ENSMUST00000113097.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr5_+_21990251 | 0.67 |
ENSMUST00000239497.2
ENSMUST00000030769.7 |
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chrM_+_7758 | 0.66 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr9_-_39405284 | 0.66 |
ENSMUST00000077757.7
ENSMUST00000215065.3 ENSMUST00000216316.3 |
Olfr44
|
olfactory receptor 44 |
| chrX_-_149879501 | 0.66 |
ENSMUST00000112683.9
ENSMUST00000026295.10 |
Tsr2
|
TSR2 20S rRNA accumulation |
| chr14_-_86986541 | 0.65 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr1_-_52271455 | 0.65 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chrY_-_1286623 | 0.64 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr18_-_64649497 | 0.64 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr17_+_33212143 | 0.64 |
ENSMUST00000087666.11
ENSMUST00000157017.2 |
Zfp952
|
zinc finger protein 952 |
| chr17_+_34423054 | 0.63 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_+_137731526 | 0.63 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr2_-_69542805 | 0.63 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr7_+_27869115 | 0.62 |
ENSMUST00000042405.8
|
Fbl
|
fibrillarin |
| chr5_-_45607554 | 0.61 |
ENSMUST00000015950.12
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr3_-_106057077 | 0.61 |
ENSMUST00000149836.2
|
Chil3
|
chitinase-like 3 |
| chr17_+_33212097 | 0.60 |
ENSMUST00000141815.3
|
Zfp952
|
zinc finger protein 952 |
| chr11_+_109540201 | 0.60 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr1_-_34882131 | 0.60 |
ENSMUST00000167518.8
ENSMUST00000047534.12 |
Fam168b
|
family with sequence similarity 168, member B |
| chr4_-_116851571 | 0.60 |
ENSMUST00000030446.15
|
Urod
|
uroporphyrinogen decarboxylase |
| chr12_-_101943134 | 0.59 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr16_+_95946591 | 0.58 |
ENSMUST00000023913.11
ENSMUST00000232832.2 ENSMUST00000233566.2 ENSMUST00000233273.2 |
Get1
|
guided entry of tail-anchored proteins factor 1 |
| chr10_+_19497740 | 0.58 |
ENSMUST00000036564.8
|
Il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr15_+_79543397 | 0.58 |
ENSMUST00000023064.9
|
Cby1
|
chibby family member 1, beta catenin antagonist |
| chr15_+_100251030 | 0.58 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr7_-_47101887 | 0.57 |
ENSMUST00000159004.2
|
Mrgpra2a
|
MAS-related GPR, member A2A |
| chr10_-_17898838 | 0.57 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr15_+_102204691 | 0.57 |
ENSMUST00000064924.6
ENSMUST00000230212.2 ENSMUST00000229050.2 ENSMUST00000231104.2 |
Espl1
|
extra spindle pole bodies 1, separase |
| chr4_-_123033721 | 0.56 |
ENSMUST00000030404.5
|
Ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr13_-_19579898 | 0.55 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr10_-_115197775 | 0.54 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr7_-_140462221 | 0.53 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr11_-_113540867 | 0.53 |
ENSMUST00000136392.8
ENSMUST00000125890.8 ENSMUST00000146031.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr10_+_61516078 | 0.53 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr3_+_51323383 | 0.53 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr7_-_30364394 | 0.53 |
ENSMUST00000019697.9
|
Haus5
|
HAUS augmin-like complex, subunit 5 |
| chr13_+_67052978 | 0.53 |
ENSMUST00000168767.9
|
Gm10767
|
predicted gene 10767 |
| chr6_-_69282389 | 0.52 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68 |
| chr8_-_117648104 | 0.52 |
ENSMUST00000128304.2
|
Cmc2
|
COX assembly mitochondrial protein 2 |
| chr7_+_28416270 | 0.51 |
ENSMUST00000108279.9
|
Fbxo17
|
F-box protein 17 |
| chr1_+_132405099 | 0.51 |
ENSMUST00000190825.7
ENSMUST00000190997.7 ENSMUST00000187505.7 ENSMUST00000027700.15 ENSMUST00000188575.7 |
Rbbp5
|
retinoblastoma binding protein 5, histone lysine methyltransferase complex subunit |
| chr6_+_58617521 | 0.51 |
ENSMUST00000145161.8
ENSMUST00000203146.3 ENSMUST00000114294.8 ENSMUST00000204948.2 |
Abcg2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr5_+_140404997 | 0.51 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr10_-_81200680 | 0.51 |
ENSMUST00000131736.8
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr2_+_118865262 | 0.50 |
ENSMUST00000028796.2
|
Rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr11_-_96807233 | 0.50 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr17_-_81372483 | 0.50 |
ENSMUST00000025093.6
|
Thumpd2
|
THUMP domain containing 2 |
| chr15_+_100052260 | 0.50 |
ENSMUST00000023768.14
|
Dip2b
|
disco interacting protein 2 homolog B |
| chr8_+_70625032 | 0.50 |
ENSMUST00000002413.15
ENSMUST00000182980.8 ENSMUST00000182365.8 |
Tmem161a
|
transmembrane protein 161A |
| chr1_+_171041539 | 0.49 |
ENSMUST00000005820.11
ENSMUST00000075469.12 ENSMUST00000155126.8 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr9_+_74959259 | 0.49 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr11_-_51541610 | 0.49 |
ENSMUST00000142721.2
ENSMUST00000156835.8 ENSMUST00000001080.16 |
N4bp3
|
NEDD4 binding protein 3 |
| chr9_-_111086528 | 0.49 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chr6_+_134617903 | 0.49 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr7_-_140461769 | 0.49 |
ENSMUST00000106048.10
ENSMUST00000147331.9 ENSMUST00000137710.2 |
Sirt3
|
sirtuin 3 |
| chr12_-_99849660 | 0.48 |
ENSMUST00000221929.2
ENSMUST00000046485.5 |
Efcab11
|
EF-hand calcium binding domain 11 |
| chr8_-_111754379 | 0.48 |
ENSMUST00000040241.15
|
Ddx19b
|
DEAD box helicase 19b |
| chr8_-_117648147 | 0.47 |
ENSMUST00000078589.7
ENSMUST00000148235.8 |
Cmc2
|
COX assembly mitochondrial protein 2 |
| chr8_-_73059104 | 0.47 |
ENSMUST00000075602.8
|
Gm10282
|
predicted pseudogene 10282 |
| chr17_+_24645615 | 0.47 |
ENSMUST00000234304.2
ENSMUST00000024946.7 ENSMUST00000234150.2 |
Eci1
|
enoyl-Coenzyme A delta isomerase 1 |
| chr2_-_93292257 | 0.46 |
ENSMUST00000150508.8
|
Cd82
|
CD82 antigen |
| chr9_-_36637923 | 0.46 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr7_+_27869192 | 0.46 |
ENSMUST00000208967.2
|
Fbl
|
fibrillarin |
| chr9_+_89081407 | 0.46 |
ENSMUST00000138109.2
|
Gm29094
|
predicted gene 29094 |
| chr4_-_46389391 | 0.45 |
ENSMUST00000086563.11
ENSMUST00000030015.6 |
Trmo
|
tRNA methyltransferase O |
| chr7_+_119701570 | 0.45 |
ENSMUST00000033210.13
|
Tmem159
|
transmembrane protein 159 |
| chr1_-_163822336 | 0.45 |
ENSMUST00000097493.10
ENSMUST00000045876.8 |
BC055324
|
cDNA sequence BC055324 |
| chr6_-_69415741 | 0.45 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr5_+_52940391 | 0.45 |
ENSMUST00000031077.13
ENSMUST00000113904.9 ENSMUST00000199840.2 |
Zcchc4
|
zinc finger, CCHC domain containing 4 |
| chr12_+_17398421 | 0.44 |
ENSMUST00000046011.12
|
Nol10
|
nucleolar protein 10 |
| chr17_-_30790804 | 0.44 |
ENSMUST00000236799.2
ENSMUST00000237048.2 |
Btbd9
|
BTB (POZ) domain containing 9 |
| chr2_-_93292708 | 0.44 |
ENSMUST00000123565.8
|
Cd82
|
CD82 antigen |
| chr9_-_21223551 | 0.44 |
ENSMUST00000003397.9
ENSMUST00000213250.2 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr15_+_9071761 | 0.43 |
ENSMUST00000189437.8
ENSMUST00000190874.8 |
Nadk2
|
NAD kinase 2, mitochondrial |
| chr8_+_117648474 | 0.43 |
ENSMUST00000034205.5
ENSMUST00000212775.2 |
Cenpn
|
centromere protein N |
| chr5_+_115061293 | 0.43 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr17_+_21876498 | 0.43 |
ENSMUST00000039726.8
|
Zfp983
|
zinc finger protein 983 |
| chr11_+_88095222 | 0.43 |
ENSMUST00000118784.8
ENSMUST00000139170.8 ENSMUST00000107915.10 ENSMUST00000144070.3 |
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr16_-_91728162 | 0.42 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr15_+_90108480 | 0.42 |
ENSMUST00000100309.3
ENSMUST00000231200.2 |
Alg10b
|
asparagine-linked glycosylation 10B (alpha-1,2-glucosyltransferase) |
| chr13_+_21363602 | 0.42 |
ENSMUST00000222544.2
|
Trim27
|
tripartite motif-containing 27 |
| chr18_-_64649620 | 0.41 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr18_+_50164043 | 0.41 |
ENSMUST00000145726.2
ENSMUST00000128377.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr4_-_115911053 | 0.41 |
ENSMUST00000030475.3
|
Nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr10_+_42554888 | 0.41 |
ENSMUST00000040718.6
|
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr11_+_103007054 | 0.41 |
ENSMUST00000053063.7
|
Hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr11_+_116744578 | 0.41 |
ENSMUST00000021173.14
|
Mfsd11
|
major facilitator superfamily domain containing 11 |
| chr13_-_8921732 | 0.40 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr14_-_47514248 | 0.40 |
ENSMUST00000187531.8
ENSMUST00000111790.2 |
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr16_-_20121108 | 0.40 |
ENSMUST00000048642.15
ENSMUST00000232036.2 |
Parl
|
presenilin associated, rhomboid-like |
| chr3_+_36120195 | 0.40 |
ENSMUST00000196648.2
|
Acad9
|
acyl-Coenzyme A dehydrogenase family, member 9 |
| chr2_-_103591271 | 0.40 |
ENSMUST00000140895.2
|
Nat10
|
N-acetyltransferase 10 |
| chr13_-_53531391 | 0.39 |
ENSMUST00000021920.8
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr2_+_30171055 | 0.39 |
ENSMUST00000143119.3
|
Gm28038
|
predicted gene, 28038 |
| chr2_+_61542038 | 0.39 |
ENSMUST00000028278.14
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr1_+_59724108 | 0.39 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr15_+_31602252 | 0.39 |
ENSMUST00000042702.7
ENSMUST00000161061.3 |
Atpsckmt
|
ATP synthase C subunit lysine N-methyltransferase |
| chr13_-_19579961 | 0.39 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr1_-_80255156 | 0.38 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr10_+_129219952 | 0.38 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr8_-_108315024 | 0.38 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr1_+_163822438 | 0.38 |
ENSMUST00000045694.14
ENSMUST00000111490.2 |
Mettl18
|
methyltransferase like 18 |
| chr17_+_15277013 | 0.37 |
ENSMUST00000228803.2
|
Ermard
|
ER membrane associated RNA degradation |
| chr13_-_45155298 | 0.37 |
ENSMUST00000220555.2
|
Dtnbp1
|
dystrobrevin binding protein 1 |
| chr11_+_120489358 | 0.37 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr19_-_46958001 | 0.36 |
ENSMUST00000235234.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr6_-_39396691 | 0.36 |
ENSMUST00000146785.8
ENSMUST00000114823.8 |
Mkrn1
|
makorin, ring finger protein, 1 |
| chr15_+_9071331 | 0.36 |
ENSMUST00000190591.10
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr11_+_23616007 | 0.36 |
ENSMUST00000058163.11
|
Pus10
|
pseudouridylate synthase 10 |
| chr5_-_5564730 | 0.36 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr7_-_45116316 | 0.36 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr5_+_45650821 | 0.35 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr11_+_88095206 | 0.35 |
ENSMUST00000024486.14
|
Mrps23
|
mitochondrial ribosomal protein S23 |
| chr2_-_152185901 | 0.35 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr14_-_104760051 | 0.35 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr19_-_4240984 | 0.35 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr13_-_92620507 | 0.34 |
ENSMUST00000040106.9
|
Fam151b
|
family with sequence similarity 151, member B |
| chr6_-_125208738 | 0.34 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chrM_+_7779 | 0.34 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr5_-_110256019 | 0.34 |
ENSMUST00000077220.14
|
Gtpbp6
|
GTP binding protein 6 (putative) |
| chr6_+_71350411 | 0.33 |
ENSMUST00000066747.14
|
Cd8a
|
CD8 antigen, alpha chain |
| chr3_+_36120128 | 0.33 |
ENSMUST00000011492.15
|
Acad9
|
acyl-Coenzyme A dehydrogenase family, member 9 |
| chr14_-_27230402 | 0.33 |
ENSMUST00000050480.8
ENSMUST00000223689.2 |
Ccdc66
|
coiled-coil domain containing 66 |
| chr11_-_62172164 | 0.33 |
ENSMUST00000072916.5
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr7_+_51537645 | 0.32 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr5_+_100666278 | 0.31 |
ENSMUST00000151414.8
|
Cops4
|
COP9 signalosome subunit 4 |
| chr18_-_42395131 | 0.31 |
ENSMUST00000236102.2
|
Lars
|
leucyl-tRNA synthetase |
| chr9_+_55057334 | 0.31 |
ENSMUST00000122441.2
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr2_-_103591522 | 0.30 |
ENSMUST00000028608.13
|
Nat10
|
N-acetyltransferase 10 |
| chr3_-_107839133 | 0.30 |
ENSMUST00000004137.11
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr2_-_174305856 | 0.30 |
ENSMUST00000016396.8
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr3_-_148696155 | 0.30 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr1_-_180971763 | 0.29 |
ENSMUST00000027797.9
|
Nvl
|
nuclear VCP-like |
| chr5_-_21990170 | 0.29 |
ENSMUST00000115193.8
ENSMUST00000115192.2 ENSMUST00000115195.8 ENSMUST00000030771.12 |
Dnajc2
|
DnaJ heat shock protein family (Hsp40) member C2 |
| chr4_-_126954878 | 0.29 |
ENSMUST00000136186.2
ENSMUST00000106099.8 ENSMUST00000106102.9 |
Zmym1
|
zinc finger, MYM domain containing 1 |
| chr5_-_149559636 | 0.29 |
ENSMUST00000201452.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr2_-_86109346 | 0.29 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr2_-_30249202 | 0.29 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr5_-_149559667 | 0.29 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_+_90128086 | 0.29 |
ENSMUST00000065418.7
|
Rab13
|
RAB13, member RAS oncogene family |
| chr15_-_36792649 | 0.29 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr9_+_19533374 | 0.29 |
ENSMUST00000213725.2
ENSMUST00000208694.2 |
Zfp317
|
zinc finger protein 317 |
| chr1_+_33758937 | 0.28 |
ENSMUST00000088287.10
|
Rab23
|
RAB23, member RAS oncogene family |
| chr6_+_71350519 | 0.28 |
ENSMUST00000172321.3
|
Cd8a
|
CD8 antigen, alpha chain |
| chr2_+_85809620 | 0.27 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr12_+_65122355 | 0.27 |
ENSMUST00000058889.5
|
Fancm
|
Fanconi anemia, complementation group M |
| chr11_+_101518768 | 0.27 |
ENSMUST00000010506.10
|
Rdm1
|
RAD52 motif 1 |
| chr18_-_70663382 | 0.27 |
ENSMUST00000043286.15
|
Poli
|
polymerase (DNA directed), iota |
| chrX_+_105059305 | 0.27 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr7_-_37927399 | 0.27 |
ENSMUST00000098513.6
|
Plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr7_-_100164007 | 0.26 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr2_-_93292734 | 0.26 |
ENSMUST00000099696.8
|
Cd82
|
CD82 antigen |
| chr5_-_149559792 | 0.25 |
ENSMUST00000202361.4
ENSMUST00000202089.4 ENSMUST00000200825.2 ENSMUST00000201559.4 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_+_4928260 | 0.25 |
ENSMUST00000165720.3
|
Tcea1
|
transcription elongation factor A (SII) 1 |
| chr5_-_8472582 | 0.24 |
ENSMUST00000168500.8
ENSMUST00000002368.16 |
Dbf4
|
DBF4 zinc finger |
| chr6_-_54949587 | 0.24 |
ENSMUST00000060655.15
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr1_+_121358778 | 0.24 |
ENSMUST00000036025.16
ENSMUST00000112621.2 |
Ccdc93
|
coiled-coil domain containing 93 |
| chr13_-_117161921 | 0.24 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr18_-_24663260 | 0.24 |
ENSMUST00000046206.5
|
Rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr1_+_78635542 | 0.24 |
ENSMUST00000035779.15
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr2_-_180928867 | 0.23 |
ENSMUST00000130475.8
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr8_-_70959360 | 0.23 |
ENSMUST00000136913.2
ENSMUST00000075175.12 |
Rex1bd
|
required for excision 1-B domain containing |
| chr7_-_46558754 | 0.23 |
ENSMUST00000209538.2
|
Tsg101
|
tumor susceptibility gene 101 |
| chrX_+_36059274 | 0.23 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr13_-_25204272 | 0.23 |
ENSMUST00000021772.4
|
Mrs2
|
MRS2 magnesium transporter |
| chr12_-_79030250 | 0.23 |
ENSMUST00000070174.14
|
Tmem229b
|
transmembrane protein 229B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Homez
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0002477 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.4 | 1.3 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.3 | 1.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 1.5 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.3 | 1.1 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.3 | 1.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.3 | 1.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.7 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.2 | 0.5 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 0.5 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.1 | 1.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.6 | GO:0061646 | positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization(GO:0061646) |
| 0.1 | 0.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 2.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.4 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.1 | 0.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 0.8 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.3 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 0.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.0 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 1.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.0 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.9 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.6 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.4 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 1.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.6 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.1 | 0.6 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.2 | GO:0006407 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.1 | 0.4 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.2 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.6 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.5 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 1.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.8 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.7 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.0 | 0.2 | GO:0046477 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.7 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.4 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.4 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.0 | GO:0042128 | nitrate assimilation(GO:0042128) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.5 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.4 | GO:0097144 | BAX complex(GO:0097144) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 1.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.2 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.0 | 0.8 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.9 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.4 | 1.3 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.3 | 1.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 1.1 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.3 | 1.0 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 0.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 0.7 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.2 | 0.5 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 0.4 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 0.5 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 1.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.2 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 1.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |