Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
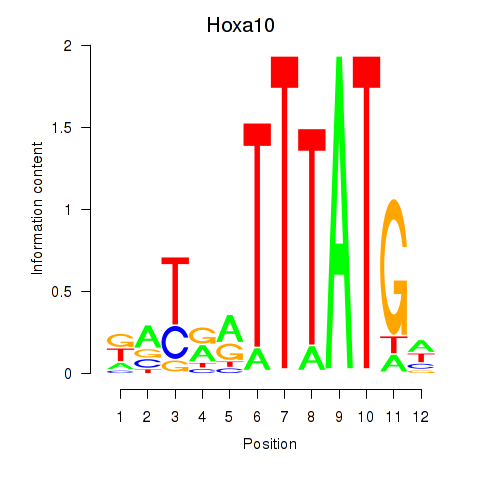
Results for Hoxa10
Z-value: 1.34
Transcription factors associated with Hoxa10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa10
|
ENSMUSG00000000938.12 | homeobox A10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa10 | mm39_v1_chr6_-_52211882_52211940 | 0.50 | 4.0e-01 | Click! |
Activity profile of Hoxa10 motif
Sorted Z-values of Hoxa10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23765546 | 2.28 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr1_-_71692320 | 1.79 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr3_+_60380243 | 1.45 |
ENSMUST00000195724.6
|
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_-_140111138 | 1.08 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 1.05 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr2_+_87610895 | 0.99 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152 |
| chr14_-_50564218 | 0.97 |
ENSMUST00000217152.2
|
Olfr734
|
olfactory receptor 734 |
| chr11_-_58614840 | 0.88 |
ENSMUST00000214728.2
|
Olfr318
|
olfactory receptor 318 |
| chrM_+_3906 | 0.80 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrM_+_14138 | 0.77 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.77 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr2_-_86528739 | 0.69 |
ENSMUST00000214141.2
|
Olfr1087
|
olfactory receptor 1087 |
| chr1_+_74752710 | 0.67 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr7_-_102774821 | 0.63 |
ENSMUST00000211075.3
ENSMUST00000215304.2 ENSMUST00000213281.2 |
Olfr586
|
olfactory receptor 586 |
| chr7_-_10292412 | 0.60 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68 |
| chr1_-_156767123 | 0.59 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_+_19404591 | 0.57 |
ENSMUST00000214130.2
|
Olfr851
|
olfactory receptor 851 |
| chr3_+_94305824 | 0.55 |
ENSMUST00000050975.6
|
Lingo4
|
leucine rich repeat and Ig domain containing 4 |
| chr10_-_129524028 | 0.53 |
ENSMUST00000203785.3
ENSMUST00000217576.2 |
Olfr802
|
olfactory receptor 802 |
| chr18_-_80751327 | 0.51 |
ENSMUST00000236310.2
ENSMUST00000167977.8 ENSMUST00000035800.8 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr7_-_104991477 | 0.49 |
ENSMUST00000213290.2
|
Olfr691
|
olfactory receptor 691 |
| chr15_+_3300249 | 0.48 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr13_-_42000958 | 0.46 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr6_-_122833109 | 0.45 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr6_-_70237939 | 0.43 |
ENSMUST00000103386.3
|
Igkv6-23
|
immunoglobulin kappa variable 6-23 |
| chr14_-_54923517 | 0.43 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr3_+_60380463 | 0.42 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr1_+_136059101 | 0.40 |
ENSMUST00000075164.11
|
Kif21b
|
kinesin family member 21B |
| chr17_-_36568728 | 0.39 |
ENSMUST00000077535.5
|
Rpp21
|
ribonuclease P 21 subunit |
| chr2_-_51039112 | 0.39 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr2_-_86070633 | 0.38 |
ENSMUST00000215607.3
|
Olfr1048
|
olfactory receptor 1048 |
| chr10_-_10958031 | 0.37 |
ENSMUST00000105561.9
ENSMUST00000044306.13 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr8_+_121262528 | 0.36 |
ENSMUST00000120493.8
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr7_-_73187369 | 0.36 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr14_-_50538979 | 0.34 |
ENSMUST00000216195.2
ENSMUST00000214372.2 ENSMUST00000214756.2 |
Olfr733
|
olfactory receptor 733 |
| chr12_+_55211069 | 0.33 |
ENSMUST00000218889.2
|
Srp54b
|
signal recognition particle 54B |
| chr3_+_138058139 | 0.32 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr13_+_83720484 | 0.31 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr7_-_86016045 | 0.31 |
ENSMUST00000213255.2
ENSMUST00000216700.2 ENSMUST00000213869.2 |
Olfr305
|
olfactory receptor 305 |
| chr16_-_58898368 | 0.30 |
ENSMUST00000216495.3
|
Olfr190
|
olfactory receptor 190 |
| chrX_+_102400061 | 0.27 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr13_-_64277115 | 0.27 |
ENSMUST00000220792.2
ENSMUST00000222866.2 ENSMUST00000099441.6 ENSMUST00000222168.2 |
Slc35d2
|
solute carrier family 35, member D2 |
| chr17_+_37689924 | 0.27 |
ENSMUST00000215518.2
|
Olfr105-ps
|
olfactory receptor 105, pseudogene |
| chr17_+_17669082 | 0.27 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr9_+_38259707 | 0.26 |
ENSMUST00000217063.2
|
Olfr898
|
olfactory receptor 898 |
| chrX_+_158242121 | 0.25 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr1_-_80642969 | 0.25 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr2_+_88644840 | 0.24 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr2_+_102488985 | 0.24 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr17_+_15720150 | 0.24 |
ENSMUST00000159197.8
ENSMUST00000014911.12 ENSMUST00000162505.8 ENSMUST00000147081.9 ENSMUST00000118001.8 ENSMUST00000143924.8 ENSMUST00000119879.9 ENSMUST00000155051.8 ENSMUST00000117593.8 |
Tbp
|
TATA box binding protein |
| chr1_-_156767196 | 0.23 |
ENSMUST00000185198.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr8_+_13034245 | 0.23 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr9_-_50650663 | 0.23 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr17_-_33979442 | 0.20 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr9_+_109865810 | 0.20 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr9_+_38432876 | 0.20 |
ENSMUST00000216496.2
|
Olfr911-ps1
|
olfactory receptor 911, pseudogene 1 |
| chr12_-_16639721 | 0.18 |
ENSMUST00000221146.2
|
Lpin1
|
lipin 1 |
| chr10_-_30647881 | 0.18 |
ENSMUST00000215740.2
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr7_-_103320398 | 0.18 |
ENSMUST00000062144.4
|
Olfr624
|
olfactory receptor 624 |
| chr3_+_66892979 | 0.18 |
ENSMUST00000162362.8
ENSMUST00000065074.14 ENSMUST00000065047.13 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_96465265 | 0.18 |
ENSMUST00000074519.13
ENSMUST00000049093.8 |
Txnip
|
thioredoxin interacting protein |
| chr10_-_30647836 | 0.17 |
ENSMUST00000215926.2
ENSMUST00000213836.2 |
Ncoa7
|
nuclear receptor coactivator 7 |
| chr15_+_38869667 | 0.17 |
ENSMUST00000022906.8
|
Fzd6
|
frizzled class receptor 6 |
| chr2_-_88600832 | 0.17 |
ENSMUST00000217588.3
|
Olfr1200
|
olfactory receptor 1200 |
| chr1_+_153750081 | 0.16 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr17_-_6129968 | 0.16 |
ENSMUST00000024570.6
ENSMUST00000097432.10 |
Serac1
|
serine active site containing 1 |
| chr2_-_88519531 | 0.16 |
ENSMUST00000213545.2
|
Olfr1195
|
olfactory receptor 1195 |
| chr3_+_101917392 | 0.16 |
ENSMUST00000196324.2
|
Nhlh2
|
nescient helix loop helix 2 |
| chr8_+_111345209 | 0.16 |
ENSMUST00000034190.11
|
Vac14
|
Vac14 homolog (S. cerevisiae) |
| chr2_-_172296662 | 0.16 |
ENSMUST00000161334.2
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr2_+_164664920 | 0.15 |
ENSMUST00000132282.2
|
Zswim1
|
zinc finger SWIM-type containing 1 |
| chr9_+_38259893 | 0.15 |
ENSMUST00000216304.2
|
Olfr898
|
olfactory receptor 898 |
| chr2_+_85876205 | 0.15 |
ENSMUST00000213496.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr11_-_110058899 | 0.14 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr6_-_70412460 | 0.14 |
ENSMUST00000103394.2
|
Igkv6-14
|
immunoglobulin kappa variable 6-14 |
| chr8_+_47439916 | 0.14 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr6_-_90758954 | 0.14 |
ENSMUST00000238821.2
|
Iqsec1
|
IQ motif and Sec7 domain 1 |
| chr1_+_74401267 | 0.14 |
ENSMUST00000097697.8
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr11_-_87783073 | 0.13 |
ENSMUST00000213672.2
ENSMUST00000213928.2 |
Olfr462
|
olfactory receptor 462 |
| chr13_-_90237179 | 0.13 |
ENSMUST00000161396.2
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr8_+_107662352 | 0.13 |
ENSMUST00000212524.2
ENSMUST00000047425.5 |
Sntb2
|
syntrophin, basic 2 |
| chr6_-_57938488 | 0.13 |
ENSMUST00000228097.2
|
Vmn1r24
|
vomeronasal 1 receptor 24 |
| chr8_-_34237752 | 0.13 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr18_+_37630044 | 0.12 |
ENSMUST00000059571.7
|
Pcdhb19
|
protocadherin beta 19 |
| chr7_+_44926925 | 0.12 |
ENSMUST00000210861.2
|
Slc6a21
|
solute carrier family 6 member 21 |
| chr5_-_29583300 | 0.12 |
ENSMUST00000196321.5
ENSMUST00000200564.5 ENSMUST00000055195.11 ENSMUST00000198105.5 ENSMUST00000179191.6 |
Lmbr1
|
limb region 1 |
| chr11_-_69686756 | 0.11 |
ENSMUST00000045971.9
|
Chrnb1
|
cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) |
| chr1_-_160079007 | 0.11 |
ENSMUST00000191909.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr5_+_104447037 | 0.11 |
ENSMUST00000031246.9
|
Ibsp
|
integrin binding sialoprotein |
| chr1_+_127657142 | 0.11 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr5_-_123804745 | 0.10 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr6_+_54794433 | 0.10 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr8_-_43981143 | 0.10 |
ENSMUST00000080135.5
|
Adam26b
|
a disintegrin and metallopeptidase domain 26B |
| chr3_+_137329433 | 0.10 |
ENSMUST00000053855.8
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr10_-_128016135 | 0.10 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr3_-_37286714 | 0.10 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr18_-_10706701 | 0.09 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr6_+_42885812 | 0.09 |
ENSMUST00000216408.2
|
Olfr447
|
olfactory receptor 447 |
| chr15_+_38869415 | 0.09 |
ENSMUST00000179165.9
|
Fzd6
|
frizzled class receptor 6 |
| chr7_+_86444235 | 0.09 |
ENSMUST00000233099.2
ENSMUST00000164996.2 |
Vmn2r77
|
vomeronasal 2, receptor 77 |
| chr13_-_74882328 | 0.09 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr7_-_144024451 | 0.08 |
ENSMUST00000033407.13
|
Cttn
|
cortactin |
| chr1_-_179572765 | 0.08 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr2_-_77776719 | 0.07 |
ENSMUST00000065889.10
|
Cwc22
|
CWC22 spliceosome-associated protein |
| chr19_+_13385213 | 0.07 |
ENSMUST00000216910.3
|
Olfr1469
|
olfactory receptor 1469 |
| chr10_+_129306867 | 0.07 |
ENSMUST00000213222.2
|
Olfr788
|
olfactory receptor 788 |
| chr10_-_129509659 | 0.07 |
ENSMUST00000213294.2
ENSMUST00000216067.3 ENSMUST00000203424.2 |
Olfr801
|
olfactory receptor 801 |
| chr3_-_108797022 | 0.06 |
ENSMUST00000180063.8
ENSMUST00000053065.8 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr12_+_25024913 | 0.06 |
ENSMUST00000066652.7
ENSMUST00000220459.2 ENSMUST00000222941.2 |
Kidins220
|
kinase D-interacting substrate 220 |
| chr13_-_42001102 | 0.06 |
ENSMUST00000121404.8
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr19_+_11943265 | 0.06 |
ENSMUST00000025590.11
|
Osbp
|
oxysterol binding protein |
| chr16_+_38182569 | 0.06 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr16_-_90866032 | 0.06 |
ENSMUST00000035689.8
ENSMUST00000114076.2 |
4932438H23Rik
|
RIKEN cDNA 4932438H23 gene |
| chr2_-_164041997 | 0.06 |
ENSMUST00000063251.3
|
Wfdc15a
|
WAP four-disulfide core domain 15A |
| chr2_+_61408875 | 0.06 |
ENSMUST00000112502.8
ENSMUST00000078074.9 |
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr2_-_86061745 | 0.06 |
ENSMUST00000216056.2
|
Olfr1047
|
olfactory receptor 1047 |
| chr6_+_70192384 | 0.06 |
ENSMUST00000103383.3
|
Igkv6-25
|
immunoglobulin kappa chain variable 6-25 |
| chr18_-_36859732 | 0.05 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr9_+_38516398 | 0.05 |
ENSMUST00000217057.2
|
Olfr914
|
olfactory receptor 914 |
| chr7_+_26134538 | 0.05 |
ENSMUST00000068767.10
|
Nlrp4a
|
NLR family, pyrin domain containing 4A |
| chr2_-_77776675 | 0.05 |
ENSMUST00000111821.9
ENSMUST00000111818.8 |
Cwc22
|
CWC22 spliceosome-associated protein |
| chr13_+_23879775 | 0.05 |
ENSMUST00000041052.5
|
H1f6
|
H1.6 linker histone, cluster member |
| chrX_+_164953444 | 0.05 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr16_+_56298228 | 0.05 |
ENSMUST00000231832.2
ENSMUST00000096013.11 ENSMUST00000048471.15 ENSMUST00000231870.2 ENSMUST00000171000.3 ENSMUST00000231781.2 ENSMUST00000096012.11 |
Abi3bp
|
ABI family member 3 binding protein |
| chr19_+_43428843 | 0.04 |
ENSMUST00000223787.2
ENSMUST00000165311.3 |
Cnnm1
|
cyclin M1 |
| chr6_-_70121150 | 0.04 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr2_-_73283010 | 0.04 |
ENSMUST00000151939.2
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr15_+_59246080 | 0.04 |
ENSMUST00000168722.3
|
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr9_+_54446268 | 0.03 |
ENSMUST00000060242.12
|
Sh2d7
|
SH2 domain containing 7 |
| chr5_-_118382926 | 0.03 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr7_-_103428905 | 0.03 |
ENSMUST00000216811.2
|
Olfr68
|
olfactory receptor 68 |
| chr12_+_71936500 | 0.03 |
ENSMUST00000221317.2
|
Daam1
|
dishevelled associated activator of morphogenesis 1 |
| chr12_-_72964646 | 0.03 |
ENSMUST00000044000.12
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr1_-_172722589 | 0.03 |
ENSMUST00000027824.7
|
Apcs
|
serum amyloid P-component |
| chr9_+_35580920 | 0.03 |
ENSMUST00000118254.2
|
Pate2
|
prostate and testis expressed 2 |
| chr4_-_109059414 | 0.03 |
ENSMUST00000160774.8
ENSMUST00000194478.6 ENSMUST00000030288.14 ENSMUST00000162787.9 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr12_-_84031622 | 0.03 |
ENSMUST00000164935.3
|
Heatr4
|
HEAT repeat containing 4 |
| chr9_+_19047613 | 0.03 |
ENSMUST00000215699.2
|
Olfr837
|
olfactory receptor 837 |
| chr11_+_51895166 | 0.03 |
ENSMUST00000109076.2
|
Cdkl3
|
cyclin-dependent kinase-like 3 |
| chr7_-_130748035 | 0.03 |
ENSMUST00000070980.4
|
4933402N03Rik
|
RIKEN cDNA 4933402N03 gene |
| chr10_+_40446326 | 0.03 |
ENSMUST00000078314.14
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16 |
| chr9_+_37765565 | 0.02 |
ENSMUST00000213956.2
|
Olfr877
|
olfactory receptor 877 |
| chr6_+_89863659 | 0.02 |
ENSMUST00000226760.2
ENSMUST00000228700.2 ENSMUST00000226171.2 ENSMUST00000227747.2 |
Vmn1r44
|
vomeronasal 1 receptor 44 |
| chr13_+_110063364 | 0.02 |
ENSMUST00000117420.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr15_+_78819119 | 0.02 |
ENSMUST00000138880.9
ENSMUST00000041164.4 |
Nol12
|
nucleolar protein 12 |
| chr1_-_127782735 | 0.02 |
ENSMUST00000208183.3
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr8_+_47439948 | 0.02 |
ENSMUST00000119686.2
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr14_+_54923655 | 0.02 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr10_+_38841511 | 0.02 |
ENSMUST00000019992.6
|
Lama4
|
laminin, alpha 4 |
| chr12_-_113896002 | 0.02 |
ENSMUST00000103463.3
|
Ighv14-1
|
immunoglobulin heavy variable 14-1 |
| chr6_+_54241830 | 0.02 |
ENSMUST00000146114.8
|
Chn2
|
chimerin 2 |
| chr1_-_82746169 | 0.02 |
ENSMUST00000027331.3
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr11_-_78875689 | 0.02 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr1_+_66360865 | 0.02 |
ENSMUST00000114013.8
|
Map2
|
microtubule-associated protein 2 |
| chr3_-_94343874 | 0.02 |
ENSMUST00000204913.3
ENSMUST00000191506.8 ENSMUST00000199678.4 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr17_-_78991691 | 0.02 |
ENSMUST00000145480.2
|
Strn
|
striatin, calmodulin binding protein |
| chr1_+_106908709 | 0.02 |
ENSMUST00000027564.8
|
Serpinb13
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr18_+_77369654 | 0.02 |
ENSMUST00000096547.11
ENSMUST00000148341.9 ENSMUST00000123410.9 |
Loxhd1
|
lipoxygenase homology domains 1 |
| chr14_-_51384236 | 0.02 |
ENSMUST00000080126.4
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr19_+_26863281 | 0.02 |
ENSMUST00000235850.2
ENSMUST00000099536.4 |
Gm815
|
predicted gene 815 |
| chr8_-_41507808 | 0.02 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr5_+_27109679 | 0.02 |
ENSMUST00000120555.8
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr17_+_6130061 | 0.02 |
ENSMUST00000039487.10
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr1_-_73055043 | 0.01 |
ENSMUST00000027374.7
|
Tnp1
|
transition protein 1 |
| chr19_+_31846154 | 0.01 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr12_-_76293459 | 0.01 |
ENSMUST00000219327.2
ENSMUST00000021453.6 ENSMUST00000218426.2 |
Tex21
|
testis expressed gene 21 |
| chr14_-_50521663 | 0.01 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr14_+_32972324 | 0.01 |
ENSMUST00000131086.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr9_-_46231282 | 0.01 |
ENSMUST00000159565.8
|
4931429L15Rik
|
RIKEN cDNA 4931429L15 gene |
| chr11_-_84404302 | 0.01 |
ENSMUST00000018841.3
|
Aatf
|
apoptosis antagonizing transcription factor |
| chr4_-_102883905 | 0.01 |
ENSMUST00000084382.6
ENSMUST00000106869.3 |
Insl5
|
insulin-like 5 |
| chr7_-_103261741 | 0.01 |
ENSMUST00000052152.3
|
Olfr620
|
olfactory receptor 620 |
| chr5_-_70999547 | 0.01 |
ENSMUST00000199705.2
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
| chr3_-_57209357 | 0.01 |
ENSMUST00000196979.5
ENSMUST00000029376.13 |
Tm4sf1
|
transmembrane 4 superfamily member 1 |
| chr1_+_165596961 | 0.01 |
ENSMUST00000040298.5
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr9_+_37766116 | 0.01 |
ENSMUST00000086063.4
|
Olfr877
|
olfactory receptor 877 |
| chr15_-_54783357 | 0.01 |
ENSMUST00000167541.3
ENSMUST00000171545.9 ENSMUST00000041591.16 ENSMUST00000173516.8 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_-_30563919 | 0.01 |
ENSMUST00000172697.8
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr3_-_92050043 | 0.01 |
ENSMUST00000197811.2
ENSMUST00000029535.6 |
4930511M18Rik
Lelp1
|
RIKEN cDNA 4930511M18 gene late cornified envelope-like proline-rich 1 |
| chr2_-_42543012 | 0.01 |
ENSMUST00000142688.2
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr11_-_12362136 | 0.01 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr5_+_146016064 | 0.01 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr4_+_114914607 | 0.01 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr4_-_44704006 | 0.01 |
ENSMUST00000146335.8
|
Pax5
|
paired box 5 |
| chr11_-_102469896 | 0.01 |
ENSMUST00000107080.2
|
Gm11627
|
predicted gene 11627 |
| chr5_-_103247920 | 0.01 |
ENSMUST00000112848.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr3_-_85909798 | 0.01 |
ENSMUST00000061343.4
|
Prss48
|
protease, serine 48 |
| chr6_-_42686970 | 0.01 |
ENSMUST00000045054.11
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr18_+_87774402 | 0.01 |
ENSMUST00000091776.7
|
Gm5096
|
predicted gene 5096 |
| chr15_-_3333003 | 0.01 |
ENSMUST00000165386.2
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr11_+_73051228 | 0.01 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr2_-_42543069 | 0.01 |
ENSMUST00000203080.3
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr6_-_23655130 | 0.01 |
ENSMUST00000104979.2
|
Rnf148
|
ring finger protein 148 |
| chr9_-_19389543 | 0.01 |
ENSMUST00000211832.2
ENSMUST00000077347.3 |
Olfr850
|
olfactory receptor 850 |
| chr12_-_113542610 | 0.01 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr7_+_86312872 | 0.01 |
ENSMUST00000081474.2
|
Olfr293
|
olfactory receptor 293 |
| chr2_-_42542938 | 0.01 |
ENSMUST00000204204.2
|
Lrp1b
|
low density lipoprotein-related protein 1B |
| chr16_-_58738861 | 0.01 |
ENSMUST00000171656.3
ENSMUST00000205883.2 |
Olfr180
|
olfactory receptor 180 |
| chr9_+_51027300 | 0.00 |
ENSMUST00000114431.4
ENSMUST00000213117.2 |
Btg4
|
BTG anti-proliferation factor 4 |
| chr14_-_52728503 | 0.00 |
ENSMUST00000073571.6
|
Olfr1507
|
olfactory receptor 1507 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 2.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.5 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.8 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.8 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) tolerance induction dependent upon immune response(GO:0002461) |
| 0.1 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.0 | 0.0 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.8 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.2 | 1.8 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 1.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.2 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |