Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
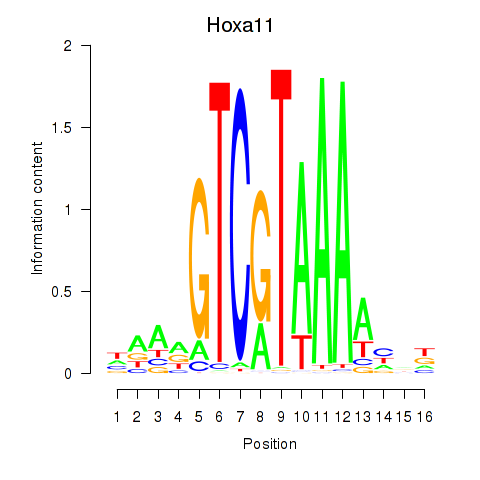
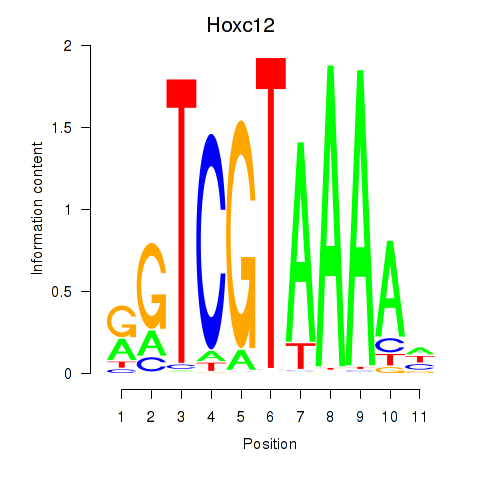
Results for Hoxa11_Hoxc12
Z-value: 0.56
Transcription factors associated with Hoxa11_Hoxc12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa11
|
ENSMUSG00000038210.11 | homeobox A11 |
|
Hoxc12
|
ENSMUSG00000050328.3 | homeobox C12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc12 | mm39_v1_chr15_+_102845192_102845269 | -0.56 | 3.2e-01 | Click! |
| Hoxa11 | mm39_v1_chr6_-_52222776_52222806 | 0.25 | 6.9e-01 | Click! |
Activity profile of Hoxa11_Hoxc12 motif
Sorted Z-values of Hoxa11_Hoxc12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_52181393 | 0.48 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr2_-_165996716 | 0.29 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr19_+_4147391 | 0.26 |
ENSMUST00000174514.2
ENSMUST00000174149.8 |
Cdk2ap2
|
CDK2-associated protein 2 |
| chr6_-_52217821 | 0.23 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr14_+_80237691 | 0.20 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr11_-_43317017 | 0.18 |
ENSMUST00000101340.11
ENSMUST00000118368.8 ENSMUST00000020685.10 ENSMUST00000020687.15 |
Pttg1
|
pituitary tumor-transforming gene 1 |
| chr16_+_13572409 | 0.15 |
ENSMUST00000239037.2
|
A930007A09Rik
|
RIKEN cDNA A930007A09 gene |
| chr1_+_178356678 | 0.14 |
ENSMUST00000161017.8
|
Kif26b
|
kinesin family member 26B |
| chr15_+_82183143 | 0.13 |
ENSMUST00000023089.5
|
Wbp2nl
|
WBP2 N-terminal like |
| chr17_+_8502682 | 0.13 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr12_-_101942463 | 0.12 |
ENSMUST00000221422.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr17_+_8502594 | 0.12 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr2_+_28095660 | 0.11 |
ENSMUST00000102879.4
ENSMUST00000028177.11 |
Olfm1
|
olfactomedin 1 |
| chr8_+_34143266 | 0.11 |
ENSMUST00000033992.9
|
Gsr
|
glutathione reductase |
| chr9_+_44318380 | 0.10 |
ENSMUST00000080300.9
|
Rps25
|
ribosomal protein S25 |
| chr9_+_44318926 | 0.10 |
ENSMUST00000216076.2
ENSMUST00000216867.2 |
Rps25
|
ribosomal protein S25 |
| chr11_-_62680273 | 0.09 |
ENSMUST00000054654.13
|
Zfp286
|
zinc finger protein 286 |
| chr6_-_52195663 | 0.09 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr9_-_50571080 | 0.09 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr9_-_106125055 | 0.07 |
ENSMUST00000074082.13
|
Alas1
|
aminolevulinic acid synthase 1 |
| chr19_+_10160884 | 0.07 |
ENSMUST00000236594.2
|
Fads1
|
fatty acid desaturase 1 |
| chr16_-_57051829 | 0.07 |
ENSMUST00000023431.8
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr4_+_132262853 | 0.07 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr1_+_87998487 | 0.06 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr13_+_98399750 | 0.06 |
ENSMUST00000022164.16
|
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr13_-_35211060 | 0.06 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr9_-_106124917 | 0.06 |
ENSMUST00000112524.9
ENSMUST00000219129.2 |
Alas1
|
aminolevulinic acid synthase 1 |
| chr7_+_126248471 | 0.06 |
ENSMUST00000032956.10
ENSMUST00000126570.8 |
Sgf29
|
SAGA complex associated factor 29 |
| chr6_+_40448334 | 0.06 |
ENSMUST00000031971.13
|
Ssbp1
|
single-stranded DNA binding protein 1 |
| chr12_-_101943134 | 0.05 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr11_-_62680228 | 0.05 |
ENSMUST00000207597.2
ENSMUST00000108705.8 |
Zfp286
|
zinc finger protein 286 |
| chr7_+_67297152 | 0.05 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_+_40448400 | 0.05 |
ENSMUST00000121360.8
ENSMUST00000117411.8 ENSMUST00000117830.2 |
Ssbp1
|
single-stranded DNA binding protein 1 |
| chr9_-_71075939 | 0.05 |
ENSMUST00000113570.8
|
Aqp9
|
aquaporin 9 |
| chr9_+_108566513 | 0.05 |
ENSMUST00000192344.2
|
Prkar2a
|
protein kinase, cAMP dependent regulatory, type II alpha |
| chr12_+_75355082 | 0.05 |
ENSMUST00000118602.8
ENSMUST00000118966.8 ENSMUST00000055390.6 |
Rhoj
|
ras homolog family member J |
| chr19_-_46327071 | 0.05 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr13_+_98399582 | 0.05 |
ENSMUST00000150352.3
ENSMUST00000226100.2 ENSMUST00000150916.9 |
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr9_+_75348800 | 0.05 |
ENSMUST00000048937.6
|
Leo1
|
Leo1, Paf1/RNA polymerase II complex component |
| chr11_-_97590460 | 0.05 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr1_+_171330978 | 0.05 |
ENSMUST00000081527.2
|
Alyref2
|
Aly/REF export factor 2 |
| chr19_-_46327024 | 0.05 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr18_-_10706701 | 0.04 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr19_-_4062738 | 0.04 |
ENSMUST00000136921.2
ENSMUST00000042497.14 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr6_-_29216299 | 0.04 |
ENSMUST00000162739.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr1_+_43769750 | 0.04 |
ENSMUST00000027217.9
|
Ecrg4
|
ECRG4 augurin precursor |
| chr6_-_22356098 | 0.04 |
ENSMUST00000165576.8
|
Fam3c
|
family with sequence similarity 3, member C |
| chr19_-_46338686 | 0.04 |
ENSMUST00000189008.3
|
2310034G01Rik
|
RIKEN cDNA 2310034G01 gene |
| chr14_-_55909314 | 0.04 |
ENSMUST00000163750.8
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr3_-_88366159 | 0.04 |
ENSMUST00000147200.8
ENSMUST00000169222.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr14_-_55909527 | 0.04 |
ENSMUST00000010520.10
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated gene 8 |
| chr11_+_115294560 | 0.04 |
ENSMUST00000153983.8
ENSMUST00000106539.10 ENSMUST00000103036.5 |
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr19_-_4062656 | 0.04 |
ENSMUST00000134479.8
ENSMUST00000128787.8 ENSMUST00000237862.2 ENSMUST00000236203.2 ENSMUST00000133474.8 |
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr3_+_89084770 | 0.04 |
ENSMUST00000029684.15
ENSMUST00000120697.8 ENSMUST00000098941.5 |
Scamp3
|
secretory carrier membrane protein 3 |
| chr17_-_22064740 | 0.04 |
ENSMUST00000084141.6
ENSMUST00000232918.2 |
Zfp820
|
zinc finger protein 820 |
| chr2_+_74542255 | 0.04 |
ENSMUST00000111983.9
|
Hoxd3
|
homeobox D3 |
| chr11_-_102588536 | 0.03 |
ENSMUST00000164506.3
ENSMUST00000092569.13 |
Ccdc43
|
coiled-coil domain containing 43 |
| chr6_+_40448286 | 0.03 |
ENSMUST00000114779.9
|
Ssbp1
|
single-stranded DNA binding protein 1 |
| chr6_+_91855015 | 0.03 |
ENSMUST00000037783.7
|
Ccdc174
|
coiled-coil domain containing 174 |
| chr17_-_13981703 | 0.03 |
ENSMUST00000127032.8
|
Tcte2
|
t-complex-associated testis expressed 2 |
| chr11_+_62349238 | 0.03 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr7_-_135130374 | 0.03 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr9_-_44318823 | 0.03 |
ENSMUST00000034623.8
|
Trappc4
|
trafficking protein particle complex 4 |
| chr8_-_70078152 | 0.03 |
ENSMUST00000121886.8
|
Zfp868
|
zinc finger protein 868 |
| chr13_+_98399693 | 0.03 |
ENSMUST00000091356.11
ENSMUST00000123924.8 |
Ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr11_+_3913970 | 0.03 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr11_-_94398162 | 0.03 |
ENSMUST00000040692.9
|
Mycbpap
|
MYCBP associated protein |
| chrX_+_55493325 | 0.03 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr16_-_57051727 | 0.03 |
ENSMUST00000226586.2
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chr9_+_62765362 | 0.02 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr6_+_149043011 | 0.02 |
ENSMUST00000179873.8
ENSMUST00000047531.16 ENSMUST00000111548.8 ENSMUST00000111547.2 ENSMUST00000134306.8 ENSMUST00000147934.4 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr1_+_182236728 | 0.02 |
ENSMUST00000117245.2
|
Trp53bp2
|
transformation related protein 53 binding protein 2 |
| chr5_-_116162415 | 0.02 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr5_-_86437119 | 0.02 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr1_+_161222980 | 0.02 |
ENSMUST00000028024.5
|
Tnfsf4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr17_+_22580434 | 0.02 |
ENSMUST00000088765.9
ENSMUST00000149699.8 ENSMUST00000072477.11 ENSMUST00000121315.2 |
Zfp758
|
zinc finger protein 758 |
| chr14_+_55909692 | 0.02 |
ENSMUST00000002397.7
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr18_-_38342815 | 0.02 |
ENSMUST00000057185.13
|
Pcdh1
|
protocadherin 1 |
| chr1_+_74752710 | 0.02 |
ENSMUST00000027356.7
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr13_-_12479804 | 0.02 |
ENSMUST00000124888.8
|
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr16_+_33614715 | 0.02 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr13_+_109822527 | 0.02 |
ENSMUST00000074103.12
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr3_+_126390951 | 0.02 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr19_-_13075176 | 0.02 |
ENSMUST00000208913.2
ENSMUST00000215229.2 |
Olfr1457
|
olfactory receptor 1457 |
| chr19_-_13074935 | 0.02 |
ENSMUST00000214561.3
|
Olfr1457
|
olfactory receptor 1457 |
| chr3_+_126391046 | 0.02 |
ENSMUST00000106401.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr1_-_74163575 | 0.02 |
ENSMUST00000169786.8
ENSMUST00000212888.2 ENSMUST00000191104.7 |
Tns1
|
tensin 1 |
| chr11_+_77107006 | 0.02 |
ENSMUST00000156488.8
ENSMUST00000037912.12 |
Ssh2
|
slingshot protein phosphatase 2 |
| chr9_+_106158212 | 0.02 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A |
| chr6_+_42263609 | 0.02 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr11_+_60370741 | 0.02 |
ENSMUST00000126522.4
|
Myo15
|
myosin XV |
| chr18_+_37466877 | 0.02 |
ENSMUST00000194655.2
ENSMUST00000061717.4 |
Pcdhb6
|
protocadherin beta 6 |
| chr14_-_29443792 | 0.02 |
ENSMUST00000022567.9
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr6_-_29216276 | 0.02 |
ENSMUST00000162215.8
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr6_+_149043136 | 0.01 |
ENSMUST00000166416.8
ENSMUST00000111551.2 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr4_+_109092459 | 0.01 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr7_-_119744509 | 0.01 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr7_+_92729067 | 0.01 |
ENSMUST00000051179.12
|
Fam181b
|
family with sequence similarity 181, member B |
| chr19_-_33602652 | 0.01 |
ENSMUST00000124230.3
|
Gm8978
|
predicted gene 8978 |
| chr11_-_99987051 | 0.01 |
ENSMUST00000103127.4
|
Krt35
|
keratin 35 |
| chr11_+_105885461 | 0.01 |
ENSMUST00000190995.2
|
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr14_+_52680447 | 0.01 |
ENSMUST00000205900.3
ENSMUST00000215030.2 ENSMUST00000206100.2 ENSMUST00000206718.2 |
Olfr1509
|
olfactory receptor 1509 |
| chr19_+_29923182 | 0.01 |
ENSMUST00000025724.9
|
Il33
|
interleukin 33 |
| chr1_-_128256048 | 0.01 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr17_-_24674252 | 0.01 |
ENSMUST00000056032.9
ENSMUST00000226941.2 |
E4f1
|
E4F transcription factor 1 |
| chr18_+_36877709 | 0.01 |
ENSMUST00000007042.6
ENSMUST00000237095.2 |
Ik
|
IK cytokine |
| chr1_+_161796854 | 0.01 |
ENSMUST00000160881.2
ENSMUST00000159648.2 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr2_+_23097482 | 0.01 |
ENSMUST00000028113.10
ENSMUST00000114505.2 |
Potegl
|
POTE ankyrin domain family, member G like |
| chr9_+_38119661 | 0.01 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893 |
| chr1_+_106908709 | 0.01 |
ENSMUST00000027564.8
|
Serpinb13
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr17_-_36983111 | 0.01 |
ENSMUST00000041662.3
|
H2-M1
|
histocompatibility 2, M region locus 1 |
| chr3_-_92627651 | 0.01 |
ENSMUST00000047153.4
|
Lce1f
|
late cornified envelope 1F |
| chr11_-_23845207 | 0.01 |
ENSMUST00000102863.3
ENSMUST00000020513.10 |
Papolg
|
poly(A) polymerase gamma |
| chr14_+_27344385 | 0.01 |
ENSMUST00000210135.2
ENSMUST00000090302.6 ENSMUST00000211087.2 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chrX_+_20230720 | 0.01 |
ENSMUST00000033372.13
ENSMUST00000115391.8 ENSMUST00000115387.8 |
Rp2
|
retinitis pigmentosa 2 homolog |
| chr11_+_59197746 | 0.01 |
ENSMUST00000000128.10
ENSMUST00000108783.4 |
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr13_-_54897660 | 0.01 |
ENSMUST00000135343.2
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1 |
| chr15_-_101759212 | 0.01 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr13_+_65660492 | 0.01 |
ENSMUST00000081471.3
|
Gm10139
|
predicted gene 10139 |
| chr16_-_19132814 | 0.01 |
ENSMUST00000216157.2
|
Olfr164
|
olfactory receptor 164 |
| chr7_-_142215595 | 0.01 |
ENSMUST00000145896.3
|
Igf2
|
insulin-like growth factor 2 |
| chr10_-_120815232 | 0.01 |
ENSMUST00000119944.8
ENSMUST00000119093.2 |
Lemd3
|
LEM domain containing 3 |
| chr19_+_58931847 | 0.01 |
ENSMUST00000054280.10
ENSMUST00000200910.4 |
Eno4
|
enolase 4 |
| chr9_+_106158549 | 0.01 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A |
| chr4_-_144291704 | 0.01 |
ENSMUST00000105748.2
|
Aadacl4fm2
|
AADACL4 family member 2 |
| chr7_-_41098120 | 0.01 |
ENSMUST00000233793.2
ENSMUST00000233555.2 ENSMUST00000165029.3 |
Vmn2r57
|
vomeronasal 2, receptor 57 |
| chr11_+_117375194 | 0.01 |
ENSMUST00000092394.4
|
Gm11733
|
predicted gene 11733 |
| chr15_+_10177709 | 0.01 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr1_-_136888118 | 0.01 |
ENSMUST00000192357.6
ENSMUST00000027649.14 |
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr17_+_75772567 | 0.01 |
ENSMUST00000234660.2
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr2_+_88015223 | 0.01 |
ENSMUST00000079599.4
|
Olfr1168
|
olfactory receptor 1168 |
| chr3_-_144555062 | 0.01 |
ENSMUST00000159989.2
|
Clca3b
|
chloride channel accessory 3B |
| chr5_+_149942140 | 0.01 |
ENSMUST00000065745.10
ENSMUST00000110496.5 ENSMUST00000201612.2 |
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr10_+_40559249 | 0.01 |
ENSMUST00000058747.4
|
Mettl24
|
methyltransferase like 24 |
| chr19_+_12647803 | 0.01 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr9_+_21231994 | 0.01 |
ENSMUST00000217461.2
|
Slc44a2
|
solute carrier family 44, member 2 |
| chr16_-_16418397 | 0.01 |
ENSMUST00000159542.8
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_88479038 | 0.01 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr2_+_20742115 | 0.01 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr2_+_22958179 | 0.01 |
ENSMUST00000227663.2
ENSMUST00000028121.15 ENSMUST00000227809.2 ENSMUST00000144088.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr2_+_87574098 | 0.01 |
ENSMUST00000214723.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr4_-_150087587 | 0.01 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr17_-_65946817 | 0.01 |
ENSMUST00000233702.2
|
Txndc2
|
thioredoxin domain containing 2 (spermatozoa) |
| chr11_+_96194333 | 0.01 |
ENSMUST00000049272.5
|
Hoxb5
|
homeobox B5 |
| chr4_+_39450265 | 0.01 |
ENSMUST00000029955.5
|
1700009N14Rik
|
RIKEN cDNA 1700009N14 gene |
| chr13_+_24023428 | 0.01 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr11_+_49321254 | 0.01 |
ENSMUST00000203369.2
|
Olfr1389
|
olfactory receptor 1389 |
| chr15_-_77854988 | 0.01 |
ENSMUST00000100484.6
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr4_-_34756222 | 0.01 |
ENSMUST00000095129.3
|
Gm136
|
predicted gene 136 |
| chr13_-_22403990 | 0.01 |
ENSMUST00000057516.2
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr6_-_55658242 | 0.01 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr13_+_23433408 | 0.01 |
ENSMUST00000091719.2
|
Vmn1r223
|
vomeronasal 1 receptor 223 |
| chr11_+_81936531 | 0.01 |
ENSMUST00000021011.3
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr7_+_107411470 | 0.01 |
ENSMUST00000208563.3
ENSMUST00000214253.2 |
Olfr467
|
olfactory receptor 467 |
| chr2_-_86088672 | 0.01 |
ENSMUST00000216185.3
|
Olfr1049
|
olfactory receptor 1049 |
| chr15_+_100321074 | 0.01 |
ENSMUST00000148928.2
|
Gm5475
|
predicted gene 5475 |
| chr18_+_42527604 | 0.01 |
ENSMUST00000025374.4
|
Pou4f3
|
POU domain, class 4, transcription factor 3 |
| chr19_+_58748132 | 0.01 |
ENSMUST00000026081.5
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr11_-_65050716 | 0.01 |
ENSMUST00000020855.5
ENSMUST00000108696.7 |
1700086D15Rik
|
RIKEN cDNA 1700086D15 gene |
| chr13_+_24023386 | 0.00 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr2_-_89774457 | 0.00 |
ENSMUST00000090695.3
|
Olfr1259
|
olfactory receptor 1259 |
| chr8_+_84016970 | 0.00 |
ENSMUST00000034146.5
|
Ucp1
|
uncoupling protein 1 (mitochondrial, proton carrier) |
| chr9_+_40785277 | 0.00 |
ENSMUST00000067375.5
|
Bsx
|
brain specific homeobox |
| chr15_-_100321973 | 0.00 |
ENSMUST00000154676.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr5_+_32768515 | 0.00 |
ENSMUST00000202543.4
ENSMUST00000072311.13 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr4_-_62069046 | 0.00 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr2_-_86086061 | 0.00 |
ENSMUST00000099899.3
|
Olfr1049
|
olfactory receptor 1049 |
| chrX_+_35664933 | 0.00 |
ENSMUST00000115253.3
|
Gm6268
|
predicted gene 6268 |
| chr7_-_107886331 | 0.00 |
ENSMUST00000210114.3
|
Olfr490
|
olfactory receptor 490 |
| chr6_+_83985495 | 0.00 |
ENSMUST00000113821.8
|
Dysf
|
dysferlin |
| chr8_+_124750133 | 0.00 |
ENSMUST00000034457.9
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr17_-_20785018 | 0.00 |
ENSMUST00000167093.3
|
Vmn2r109
|
vomeronasal 2, receptor 109 |
| chr10_+_93390740 | 0.00 |
ENSMUST00000132214.8
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr8_+_94954280 | 0.00 |
ENSMUST00000109547.2
|
Nup93
|
nucleoporin 93 |
| chr4_+_144340236 | 0.00 |
ENSMUST00000094510.4
|
Aadacl4
|
arylacetamide deacetylase like 4 |
| chr8_+_11890474 | 0.00 |
ENSMUST00000033909.14
ENSMUST00000209692.2 |
Tex29
|
testis expressed 29 |
| chr2_-_89980380 | 0.00 |
ENSMUST00000099754.5
|
Olfr1270
|
olfactory receptor 1270 |
| chr9_+_53678801 | 0.00 |
ENSMUST00000048670.10
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_-_96673423 | 0.00 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr18_-_43526411 | 0.00 |
ENSMUST00000025379.14
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr2_+_87576198 | 0.00 |
ENSMUST00000217572.2
|
Olfr1140
|
olfactory receptor 1140 |
| chr17_+_34636321 | 0.00 |
ENSMUST00000142317.8
|
BC051142
|
cDNA sequence BC051142 |
| chr13_-_63712349 | 0.00 |
ENSMUST00000192155.6
|
Ptch1
|
patched 1 |
| chr18_+_65276629 | 0.00 |
ENSMUST00000235310.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr10_+_89906956 | 0.00 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr16_+_65320163 | 0.00 |
ENSMUST00000184525.2
|
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr4_+_109092829 | 0.00 |
ENSMUST00000030285.8
|
Calr4
|
calreticulin 4 |
| chr11_-_4390745 | 0.00 |
ENSMUST00000109948.8
|
Hormad2
|
HORMA domain containing 2 |
| chr6_+_42263644 | 0.00 |
ENSMUST00000163936.8
|
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr2_+_111327525 | 0.00 |
ENSMUST00000121345.4
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
| chr2_+_166647426 | 0.00 |
ENSMUST00000099078.10
|
Arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr4_-_138095277 | 0.00 |
ENSMUST00000030535.4
|
Cda
|
cytidine deaminase |
| chrX_+_132809166 | 0.00 |
ENSMUST00000033606.15
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr16_-_19349668 | 0.00 |
ENSMUST00000078554.2
|
Olfr168
|
olfactory receptor 168 |
| chr6_-_68784692 | 0.00 |
ENSMUST00000103334.4
|
Igkv4-90
|
immunoglobulin kappa chain variable 4-90 |
| chr5_+_67126304 | 0.00 |
ENSMUST00000132991.5
|
Limch1
|
LIM and calponin homology domains 1 |
| chr17_-_20455134 | 0.00 |
ENSMUST00000167382.2
|
Vmn2r105
|
vomeronasal 2, receptor 105 |
| chr9_+_19351562 | 0.00 |
ENSMUST00000217273.2
|
Olfr849
|
olfactory receptor 849 |
| chr7_+_139827152 | 0.00 |
ENSMUST00000164583.8
ENSMUST00000093984.3 |
Scart2
|
scavenger receptor family member expressed on T cells 2 |
| chr2_-_130471891 | 0.00 |
ENSMUST00000110262.3
ENSMUST00000028761.5 |
Fastkd5
Ubox5
|
FAST kinase domains 5 U box domain containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa11_Hoxc12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060435 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.2 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.0 | 0.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |