Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxa3
Z-value: 0.22
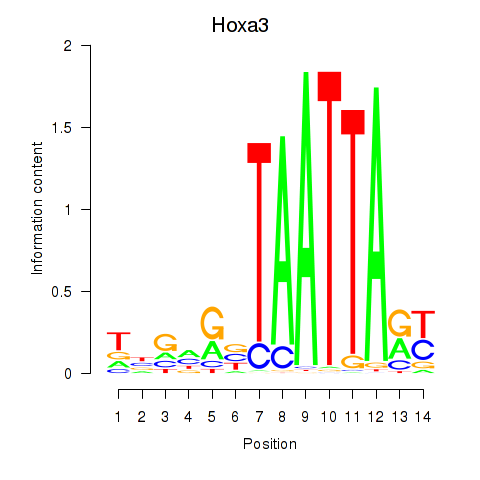
Transcription factors associated with Hoxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa3
|
ENSMUSG00000079560.14 | homeobox A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa3 | mm39_v1_chr6_-_52160816_52160837 | -0.40 | 5.0e-01 | Click! |
Activity profile of Hoxa3 motif
Sorted Z-values of Hoxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_37014859 | 0.15 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr5_-_65855511 | 0.13 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr14_+_79753055 | 0.12 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr2_+_109747984 | 0.11 |
ENSMUST00000046548.14
ENSMUST00000111037.3 |
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr2_+_36120438 | 0.11 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_+_115373895 | 0.09 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr12_+_59060162 | 0.08 |
ENSMUST00000021379.8
|
Gemin2
|
gem nuclear organelle associated protein 2 |
| chr2_-_69542805 | 0.08 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr16_-_92196954 | 0.07 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr1_-_171854818 | 0.06 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr3_+_159545309 | 0.06 |
ENSMUST00000068952.10
ENSMUST00000198878.2 |
Wls
|
wntless WNT ligand secretion mediator |
| chr15_+_65682066 | 0.06 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chrX_-_74460137 | 0.06 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr11_-_107228382 | 0.06 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr5_-_138185438 | 0.05 |
ENSMUST00000110937.8
ENSMUST00000139276.2 ENSMUST00000048698.14 ENSMUST00000123415.8 |
Taf6
|
TATA-box binding protein associated factor 6 |
| chr16_+_35861554 | 0.05 |
ENSMUST00000042203.10
|
Wdr5b
|
WD repeat domain 5B |
| chr13_-_43634695 | 0.04 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr2_+_22959452 | 0.04 |
ENSMUST00000155602.4
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr2_+_22959223 | 0.04 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr4_-_43710231 | 0.03 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr2_-_86109346 | 0.03 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr9_-_99302205 | 0.03 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr15_-_34356567 | 0.03 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr14_+_65612788 | 0.03 |
ENSMUST00000224687.2
|
Zfp395
|
zinc finger protein 395 |
| chr5_-_138185686 | 0.03 |
ENSMUST00000110936.8
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr14_+_26722319 | 0.02 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr6_-_41752111 | 0.02 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr17_-_71160477 | 0.02 |
ENSMUST00000118283.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr6_+_125529911 | 0.02 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr6_-_129449739 | 0.02 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr17_+_38104420 | 0.02 |
ENSMUST00000216051.3
|
Olfr123
|
olfactory receptor 123 |
| chr2_-_17465410 | 0.02 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr17_+_38104635 | 0.01 |
ENSMUST00000214770.3
|
Olfr123
|
olfactory receptor 123 |
| chr2_+_110551685 | 0.01 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr2_+_110551927 | 0.01 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr2_-_86088672 | 0.01 |
ENSMUST00000216185.3
|
Olfr1049
|
olfactory receptor 1049 |
| chr2_+_89642395 | 0.01 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr15_-_101801351 | 0.01 |
ENSMUST00000100179.2
|
Krt76
|
keratin 76 |
| chr13_-_103042554 | 0.01 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_136758716 | 0.01 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr7_+_107411470 | 0.00 |
ENSMUST00000208563.3
ENSMUST00000214253.2 |
Olfr467
|
olfactory receptor 467 |
| chr5_+_108842294 | 0.00 |
ENSMUST00000013633.12
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr4_-_14621805 | 0.00 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr5_+_90708962 | 0.00 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr2_+_110551976 | 0.00 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr16_-_29363671 | 0.00 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr4_-_14621497 | 0.00 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr14_-_36820304 | 0.00 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr17_+_17622934 | 0.00 |
ENSMUST00000115576.3
|
Lix1
|
limb and CNS expressed 1 |
| chr10_+_73782857 | 0.00 |
ENSMUST00000191709.6
ENSMUST00000193739.6 ENSMUST00000195531.6 |
Pcdh15
|
protocadherin 15 |
| chr10_-_129000620 | 0.00 |
ENSMUST00000214271.2
|
Olfr771
|
olfactory receptor 771 |
| chr19_-_7943365 | 0.00 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr13_-_103042294 | 0.00 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_-_21847556 | 0.00 |
ENSMUST00000214321.2
|
Olfr1361
|
olfactory receptor 1361 |
| chr2_+_88505972 | 0.00 |
ENSMUST00000216767.2
ENSMUST00000213893.2 |
Olfr1193
|
olfactory receptor 1193 |
| chr1_-_173161069 | 0.00 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chrX_-_142716085 | 0.00 |
ENSMUST00000087313.10
|
Dcx
|
doublecortin |
| chr5_-_86437119 | 0.00 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr3_-_72875187 | 0.00 |
ENSMUST00000167334.8
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr11_-_100653754 | 0.00 |
ENSMUST00000107360.3
ENSMUST00000055083.4 |
Hcrt
|
hypocretin |
| chr13_-_21798192 | 0.00 |
ENSMUST00000051874.6
|
Olfr1362
|
olfactory receptor 1362 |
| chr6_-_30936013 | 0.00 |
ENSMUST00000101589.5
|
Klf14
|
Kruppel-like factor 14 |
| chr11_-_99979052 | 0.00 |
ENSMUST00000107419.2
|
Krt32
|
keratin 32 |
| chr5_-_66672158 | 0.00 |
ENSMUST00000161879.8
ENSMUST00000159357.8 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr1_-_173018204 | 0.00 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr7_-_115637970 | 0.00 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr14_-_52648384 | 0.00 |
ENSMUST00000079459.4
|
Olfr1510
|
olfactory receptor 1510 |
| chr6_+_8948608 | 0.00 |
ENSMUST00000160300.2
|
Nxph1
|
neurexophilin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060995 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |