Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
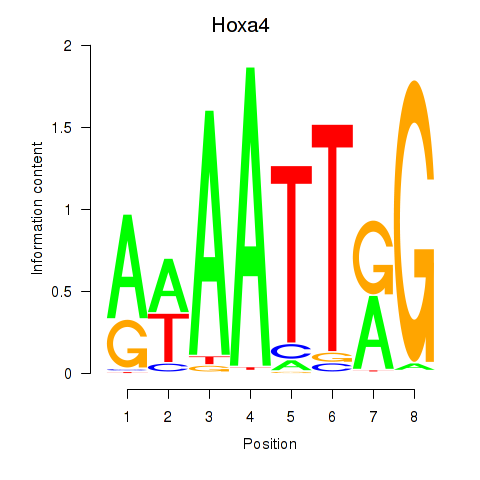
Results for Hoxa4
Z-value: 0.36
Transcription factors associated with Hoxa4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa4
|
ENSMUSG00000000942.11 | homeobox A4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa4 | mm39_v1_chr6_-_52168675_52168733 | 0.34 | 5.8e-01 | Click! |
Activity profile of Hoxa4 motif
Sorted Z-values of Hoxa4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_19432933 | 0.36 |
ENSMUST00000174355.8
ENSMUST00000172983.8 ENSMUST00000174710.2 ENSMUST00000003066.16 ENSMUST00000174064.9 |
Apoe
|
apolipoprotein E |
| chr13_-_22016364 | 0.21 |
ENSMUST00000102979.2
|
H4c18
|
H4 clustered histone 18 |
| chr9_-_114811807 | 0.12 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr6_-_52190299 | 0.12 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr9_-_49710058 | 0.11 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr2_-_5680801 | 0.11 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chrX_+_158623460 | 0.10 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr19_+_8817883 | 0.10 |
ENSMUST00000086058.13
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_+_43978377 | 0.10 |
ENSMUST00000233627.2
ENSMUST00000233437.2 |
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr5_+_42225303 | 0.10 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr8_-_69541852 | 0.09 |
ENSMUST00000037478.13
ENSMUST00000148856.2 |
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1 |
| chr9_-_49710190 | 0.09 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr19_-_23425757 | 0.09 |
ENSMUST00000036069.8
|
Mamdc2
|
MAM domain containing 2 |
| chr17_+_43978280 | 0.09 |
ENSMUST00000170988.2
|
Cyp39a1
|
cytochrome P450, family 39, subfamily a, polypeptide 1 |
| chr6_-_52168675 | 0.08 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr11_+_108814007 | 0.08 |
ENSMUST00000106711.2
|
Axin2
|
axin 2 |
| chr4_-_52919172 | 0.08 |
ENSMUST00000107667.3
ENSMUST00000213989.2 |
Olfr272
|
olfactory receptor 272 |
| chr5_+_115567644 | 0.08 |
ENSMUST00000150779.8
|
Msi1
|
musashi RNA-binding protein 1 |
| chrM_+_5319 | 0.08 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chrX_-_9335525 | 0.08 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr8_+_88978600 | 0.08 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B |
| chrM_+_10167 | 0.07 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr9_+_77543776 | 0.07 |
ENSMUST00000057781.8
|
Klhl31
|
kelch-like 31 |
| chr1_-_69723316 | 0.07 |
ENSMUST00000190855.7
ENSMUST00000188110.7 ENSMUST00000191262.7 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr16_-_76170714 | 0.07 |
ENSMUST00000231585.2
ENSMUST00000121927.8 |
Nrip1
|
nuclear receptor interacting protein 1 |
| chr5_-_124325213 | 0.07 |
ENSMUST00000161938.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr1_+_74324089 | 0.07 |
ENSMUST00000113805.8
ENSMUST00000027370.13 ENSMUST00000087226.11 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr9_-_32452885 | 0.07 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr19_-_60862964 | 0.07 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr10_+_127256736 | 0.06 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr10_-_128361731 | 0.06 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr18_+_7905440 | 0.06 |
ENSMUST00000170854.2
|
Wac
|
WW domain containing adaptor with coiled-coil |
| chr11_-_30218167 | 0.06 |
ENSMUST00000006629.14
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr2_-_13276205 | 0.06 |
ENSMUST00000191959.6
ENSMUST00000028059.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr2_-_13276074 | 0.06 |
ENSMUST00000137670.3
ENSMUST00000114791.9 |
Rsu1
|
Ras suppressor protein 1 |
| chr14_+_26722319 | 0.05 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr7_-_84339156 | 0.05 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_+_178356678 | 0.05 |
ENSMUST00000161017.8
|
Kif26b
|
kinesin family member 26B |
| chr13_-_74882374 | 0.05 |
ENSMUST00000220738.2
|
Cast
|
calpastatin |
| chr7_-_19684654 | 0.05 |
ENSMUST00000043440.8
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr10_+_129601351 | 0.05 |
ENSMUST00000203236.3
|
Olfr808
|
olfactory receptor 808 |
| chrX_-_37341274 | 0.05 |
ENSMUST00000089056.10
ENSMUST00000089054.11 ENSMUST00000066498.8 |
Tmem255a
|
transmembrane protein 255A |
| chr2_+_174169539 | 0.04 |
ENSMUST00000133356.8
ENSMUST00000087871.11 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr19_-_20931566 | 0.04 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr13_-_74882328 | 0.04 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr12_+_59176506 | 0.04 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr17_+_34457868 | 0.04 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr7_-_100504610 | 0.04 |
ENSMUST00000156855.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr10_-_126956991 | 0.04 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr1_+_87998487 | 0.04 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr8_+_12999394 | 0.04 |
ENSMUST00000110867.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr2_+_174169492 | 0.04 |
ENSMUST00000156623.8
ENSMUST00000149016.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr3_-_146388165 | 0.04 |
ENSMUST00000124931.8
ENSMUST00000147113.2 |
Samd13
|
sterile alpha motif domain containing 13 |
| chr8_+_12999480 | 0.04 |
ENSMUST00000110866.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr6_+_58810674 | 0.04 |
ENSMUST00000041401.11
|
Herc3
|
hect domain and RLD 3 |
| chr11_+_94218810 | 0.04 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr2_-_111779785 | 0.04 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chr19_-_5660057 | 0.04 |
ENSMUST00000236229.2
ENSMUST00000235701.2 ENSMUST00000236264.2 |
Kat5
|
K(lysine) acetyltransferase 5 |
| chr16_+_35891991 | 0.03 |
ENSMUST00000164916.9
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr5_-_100521343 | 0.03 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr10_+_127256993 | 0.03 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr11_+_94219046 | 0.03 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr14_+_47069667 | 0.03 |
ENSMUST00000140114.3
ENSMUST00000133989.8 |
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr3_+_107186309 | 0.03 |
ENSMUST00000199317.2
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr5_-_137529251 | 0.03 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr14_-_30213408 | 0.03 |
ENSMUST00000112250.6
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr7_-_84339045 | 0.03 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr4_+_86493905 | 0.03 |
ENSMUST00000091064.8
|
Rraga
|
Ras-related GTP binding A |
| chr13_+_22656093 | 0.03 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr2_-_62404195 | 0.03 |
ENSMUST00000174234.8
ENSMUST00000000402.16 ENSMUST00000174448.8 ENSMUST00000102732.10 |
Fap
|
fibroblast activation protein |
| chr13_+_23317325 | 0.03 |
ENSMUST00000227050.2
ENSMUST00000227160.2 ENSMUST00000227741.2 ENSMUST00000226692.2 |
Vmn1r218
|
vomeronasal 1 receptor 218 |
| chr19_+_24853039 | 0.03 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr6_-_116084810 | 0.03 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr6_+_54794433 | 0.03 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr6_+_29467717 | 0.03 |
ENSMUST00000004396.13
|
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr15_-_77840856 | 0.02 |
ENSMUST00000117725.2
ENSMUST00000016696.13 |
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr11_-_98620200 | 0.02 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr1_-_92423800 | 0.02 |
ENSMUST00000204766.2
ENSMUST00000204009.3 |
Olfr1415
|
olfactory receptor 1415 |
| chr10_+_34265969 | 0.02 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr15_-_99717956 | 0.02 |
ENSMUST00000109024.9
|
Lima1
|
LIM domain and actin binding 1 |
| chr13_+_58955506 | 0.02 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr1_+_93406686 | 0.02 |
ENSMUST00000027495.15
ENSMUST00000136182.8 ENSMUST00000131175.9 ENSMUST00000153826.8 ENSMUST00000129211.8 ENSMUST00000179353.8 ENSMUST00000172165.8 ENSMUST00000168776.8 |
Septin2
Septin2
|
septin 2 septin 2 |
| chr3_-_130524024 | 0.02 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr15_+_55171138 | 0.02 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr1_+_93406809 | 0.02 |
ENSMUST00000112912.8
|
Septin2
|
septin 2 |
| chr7_+_65343156 | 0.02 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr11_+_49313894 | 0.02 |
ENSMUST00000216641.2
ENSMUST00000217595.2 |
Olfr1389
|
olfactory receptor 1389 |
| chr17_+_80614795 | 0.02 |
ENSMUST00000223878.2
ENSMUST00000068175.6 ENSMUST00000224391.2 |
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr16_-_85698679 | 0.02 |
ENSMUST00000023611.7
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2) |
| chr18_-_15196612 | 0.02 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr8_+_4288815 | 0.02 |
ENSMUST00000003027.14
ENSMUST00000110999.8 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr9_-_114219685 | 0.02 |
ENSMUST00000084881.5
|
Crtap
|
cartilage associated protein |
| chr2_-_77349909 | 0.02 |
ENSMUST00000111830.9
|
Zfp385b
|
zinc finger protein 385B |
| chr6_-_39787813 | 0.02 |
ENSMUST00000114797.2
ENSMUST00000031978.9 |
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr12_+_85043083 | 0.02 |
ENSMUST00000168977.8
ENSMUST00000021670.15 |
Ylpm1
|
YLP motif containing 1 |
| chr11_-_99265721 | 0.02 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr7_+_131161951 | 0.02 |
ENSMUST00000084502.7
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr7_+_78922947 | 0.02 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr9_+_74959259 | 0.02 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr7_+_104263162 | 0.02 |
ENSMUST00000215575.3
|
Olfr656
|
olfactory receptor 656 |
| chr16_-_50252703 | 0.02 |
ENSMUST00000066037.13
ENSMUST00000089399.11 ENSMUST00000089404.10 ENSMUST00000138166.8 |
Bbx
|
bobby sox HMG box containing |
| chr19_-_28945194 | 0.02 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr11_+_51989508 | 0.02 |
ENSMUST00000020608.3
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform |
| chr1_-_180021039 | 0.02 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr5_+_30972067 | 0.02 |
ENSMUST00000200692.4
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr2_-_6889685 | 0.02 |
ENSMUST00000183091.8
ENSMUST00000182851.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr14_+_47069582 | 0.02 |
ENSMUST00000068532.10
|
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr13_+_38010879 | 0.02 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr4_+_133795740 | 0.02 |
ENSMUST00000121391.8
|
Crybg2
|
crystallin beta-gamma domain containing 2 |
| chr18_+_37622518 | 0.02 |
ENSMUST00000055949.4
|
Pcdhb18
|
protocadherin beta 18 |
| chr7_-_126859790 | 0.02 |
ENSMUST00000035276.5
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr3_+_95071617 | 0.02 |
ENSMUST00000168321.8
ENSMUST00000107217.6 ENSMUST00000202315.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr7_+_131162439 | 0.02 |
ENSMUST00000207442.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr12_+_59176543 | 0.02 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr4_-_62438122 | 0.02 |
ENSMUST00000107444.8
ENSMUST00000030090.4 |
Alad
|
aminolevulinate, delta-, dehydratase |
| chr15_+_98953044 | 0.01 |
ENSMUST00000230021.2
ENSMUST00000047104.15 ENSMUST00000024249.5 |
Prph
|
peripherin |
| chr8_+_4288733 | 0.01 |
ENSMUST00000110998.9
ENSMUST00000062686.11 |
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr6_+_63232955 | 0.01 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr18_+_23885390 | 0.01 |
ENSMUST00000170802.8
ENSMUST00000155708.8 ENSMUST00000118826.9 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr8_-_68427217 | 0.01 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr3_-_32546380 | 0.01 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr5_+_124065481 | 0.01 |
ENSMUST00000166129.5
|
Gm43518
|
predicted gene 43518 |
| chr16_-_96971905 | 0.01 |
ENSMUST00000056102.9
|
Dscam
|
DS cell adhesion molecule |
| chr7_+_139753130 | 0.01 |
ENSMUST00000209314.3
ENSMUST00000213953.2 ENSMUST00000214272.2 ENSMUST00000215785.2 ENSMUST00000216023.2 |
Olfr523
|
olfactory receptor 523 |
| chr1_+_133109059 | 0.01 |
ENSMUST00000187285.7
|
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr1_+_53100796 | 0.01 |
ENSMUST00000027269.7
ENSMUST00000191197.2 |
Mstn
|
myostatin |
| chr13_-_103911092 | 0.01 |
ENSMUST00000074616.7
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr18_+_37488174 | 0.01 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr7_-_109330915 | 0.01 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chr11_+_29413734 | 0.01 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr13_-_103042554 | 0.01 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_93349637 | 0.01 |
ENSMUST00000064257.6
|
Tchh
|
trichohyalin |
| chr12_-_80807454 | 0.01 |
ENSMUST00000073251.8
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr13_-_3968157 | 0.01 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr10_+_127257077 | 0.01 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr13_+_112454981 | 0.01 |
ENSMUST00000223871.2
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr9_-_37627519 | 0.01 |
ENSMUST00000215727.2
ENSMUST00000211952.3 |
Olfr160
|
olfactory receptor 160 |
| chr6_-_92511836 | 0.01 |
ENSMUST00000113446.8
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr5_+_64969679 | 0.01 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr2_+_36955758 | 0.01 |
ENSMUST00000216706.3
|
Olfr360
|
olfactory receptor 360 |
| chr19_+_31846154 | 0.01 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr12_+_84408742 | 0.01 |
ENSMUST00000021661.13
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr18_+_37440497 | 0.01 |
ENSMUST00000056712.4
|
Pcdhb4
|
protocadherin beta 4 |
| chr8_+_114932312 | 0.01 |
ENSMUST00000049509.7
ENSMUST00000150963.2 |
Vat1l
|
vesicle amine transport protein 1 like |
| chr16_+_37400500 | 0.01 |
ENSMUST00000160847.2
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr1_-_67077906 | 0.01 |
ENSMUST00000119559.8
ENSMUST00000149996.2 ENSMUST00000027149.12 ENSMUST00000113979.10 |
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1 |
| chr3_-_51184895 | 0.01 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr4_+_101407608 | 0.01 |
ENSMUST00000094953.11
ENSMUST00000106933.2 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr13_-_35100960 | 0.01 |
ENSMUST00000225242.2
|
Fam217a
|
family with sequence similarity 217, member A |
| chr14_+_65504067 | 0.01 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr7_-_10347586 | 0.01 |
ENSMUST00000226190.2
ENSMUST00000226228.2 ENSMUST00000228296.2 ENSMUST00000228638.2 ENSMUST00000176707.3 ENSMUST00000176284.9 ENSMUST00000226160.2 ENSMUST00000227853.2 ENSMUST00000228478.2 |
Vmn1r69
|
vomeronasal 1 receptor 69 |
| chr18_+_65158873 | 0.01 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr6_+_106095726 | 0.01 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr3_-_122910662 | 0.01 |
ENSMUST00000051443.8
|
Synpo2
|
synaptopodin 2 |
| chr12_-_46863726 | 0.01 |
ENSMUST00000219330.2
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr5_-_87847268 | 0.01 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr2_+_83786742 | 0.01 |
ENSMUST00000178325.2
|
Gm13698
|
predicted gene 13698 |
| chr4_+_5724305 | 0.01 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr6_+_132779781 | 0.01 |
ENSMUST00000068302.4
|
Tas2r117
|
taste receptor, type 2, member 117 |
| chr10_+_69370038 | 0.01 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr2_+_88012224 | 0.01 |
ENSMUST00000215996.2
|
Olfr1168
|
olfactory receptor 1168 |
| chr13_-_21847556 | 0.01 |
ENSMUST00000214321.2
|
Olfr1361
|
olfactory receptor 1361 |
| chr16_+_37400590 | 0.01 |
ENSMUST00000159787.8
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr13_+_111391544 | 0.01 |
ENSMUST00000054716.4
|
Actbl2
|
actin, beta-like 2 |
| chr7_+_144392323 | 0.01 |
ENSMUST00000105898.2
|
Fgf3
|
fibroblast growth factor 3 |
| chr11_+_96194333 | 0.01 |
ENSMUST00000049272.5
|
Hoxb5
|
homeobox B5 |
| chr11_+_67061908 | 0.01 |
ENSMUST00000018641.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr2_-_115896279 | 0.01 |
ENSMUST00000110907.8
ENSMUST00000110908.9 |
Meis2
|
Meis homeobox 2 |
| chr7_-_110462446 | 0.01 |
ENSMUST00000033050.5
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr1_+_172383499 | 0.01 |
ENSMUST00000061835.10
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr18_-_20247824 | 0.01 |
ENSMUST00000038710.6
|
Dsc1
|
desmocollin 1 |
| chr16_-_63684477 | 0.01 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr3_-_151953894 | 0.01 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chr6_-_138404076 | 0.01 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr3_-_64415106 | 0.01 |
ENSMUST00000170270.3
|
Vmn2r5
|
vomeronasal 2, receptor 5 |
| chr2_+_111607774 | 0.01 |
ENSMUST00000214708.2
ENSMUST00000215244.2 |
Olfr1302
|
olfactory receptor 1302 |
| chr11_+_67061837 | 0.01 |
ENSMUST00000170159.8
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult |
| chr18_+_44403169 | 0.01 |
ENSMUST00000042747.4
|
Npy6r
|
neuropeptide Y receptor Y6 |
| chr10_-_70491764 | 0.01 |
ENSMUST00000162144.2
ENSMUST00000162793.8 |
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like |
| chr13_+_58956077 | 0.01 |
ENSMUST00000109838.10
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr11_-_73483834 | 0.01 |
ENSMUST00000215689.2
|
Olfr385
|
olfactory receptor 385 |
| chr1_-_14374794 | 0.01 |
ENSMUST00000190337.7
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr4_+_114678919 | 0.01 |
ENSMUST00000137570.2
|
Gm12830
|
predicted gene 12830 |
| chr18_-_20247666 | 0.01 |
ENSMUST00000224432.2
|
Dsc1
|
desmocollin 1 |
| chr1_+_19279178 | 0.01 |
ENSMUST00000187754.7
|
Tfap2b
|
transcription factor AP-2 beta |
| chr7_+_90075762 | 0.01 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr17_+_93506435 | 0.01 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr6_+_42788873 | 0.01 |
ENSMUST00000218832.2
|
Olfr450
|
olfactory receptor 450 |
| chr17_-_36008863 | 0.01 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr2_+_70393782 | 0.01 |
ENSMUST00000123330.3
|
Gad1
|
glutamate decarboxylase 1 |
| chr6_+_40941688 | 0.01 |
ENSMUST00000076638.7
|
1810009J06Rik
|
RIKEN cDNA 1810009J06 gene |
| chr19_-_34231600 | 0.01 |
ENSMUST00000238147.2
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr7_-_119744509 | 0.01 |
ENSMUST00000208874.2
ENSMUST00000033207.6 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr6_-_12749409 | 0.01 |
ENSMUST00000119581.7
|
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr18_+_37568647 | 0.01 |
ENSMUST00000055495.6
|
Pcdhb12
|
protocadherin beta 12 |
| chr5_-_122140403 | 0.01 |
ENSMUST00000134326.2
|
Cux2
|
cut-like homeobox 2 |
| chr4_-_3872105 | 0.01 |
ENSMUST00000105158.2
|
Mos
|
Moloney sarcoma oncogene |
| chr2_+_170573727 | 0.01 |
ENSMUST00000029075.5
|
Dok5
|
docking protein 5 |
| chr17_+_93506590 | 0.01 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr12_+_103524690 | 0.01 |
ENSMUST00000187155.7
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.1 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.1 | 0.2 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |