Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Hoxa6
Z-value: 0.33
Transcription factors associated with Hoxa6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa6
|
ENSMUSG00000043219.10 | homeobox A6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa6 | mm39_v1_chr6_-_52185674_52185702 | 0.12 | 8.4e-01 | Click! |
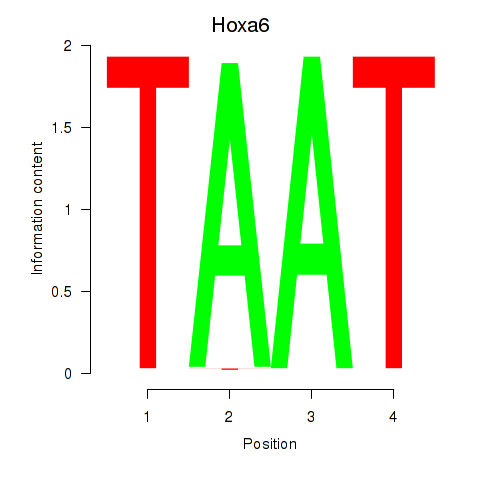
Activity profile of Hoxa6 motif
Sorted Z-values of Hoxa6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_94954202 | 0.70 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr9_+_120758282 | 0.33 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr13_-_29137673 | 0.31 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr7_-_108769719 | 0.28 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr17_-_24292453 | 0.28 |
ENSMUST00000017090.6
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr3_+_41517790 | 0.25 |
ENSMUST00000163764.8
|
Jade1
|
jade family PHD finger 1 |
| chr10_-_35587888 | 0.25 |
ENSMUST00000080898.4
|
Amd2
|
S-adenosylmethionine decarboxylase 2 |
| chr7_-_108774367 | 0.24 |
ENSMUST00000207178.2
|
Lmo1
|
LIM domain only 1 |
| chr14_+_26722319 | 0.24 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr3_+_41517829 | 0.23 |
ENSMUST00000170711.8
|
Jade1
|
jade family PHD finger 1 |
| chrM_+_8603 | 0.23 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr10_-_40178182 | 0.21 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr10_+_99099084 | 0.20 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr18_+_69633741 | 0.20 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr2_+_132689640 | 0.19 |
ENSMUST00000124836.8
ENSMUST00000154160.2 |
Crls1
|
cardiolipin synthase 1 |
| chr1_-_165762469 | 0.19 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr12_-_73093953 | 0.18 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr7_-_112946481 | 0.17 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr6_-_52203146 | 0.15 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr19_-_24178000 | 0.15 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr4_-_87951565 | 0.15 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr16_-_92196954 | 0.14 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr13_+_83652352 | 0.14 |
ENSMUST00000198916.5
ENSMUST00000200123.5 ENSMUST00000005722.14 ENSMUST00000163888.8 |
Mef2c
|
myocyte enhancer factor 2C |
| chrM_+_7758 | 0.14 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr6_+_29859372 | 0.14 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr9_+_72600721 | 0.14 |
ENSMUST00000238315.2
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4 |
| chr19_+_26727111 | 0.13 |
ENSMUST00000175842.4
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_-_105398346 | 0.12 |
ENSMUST00000176770.8
ENSMUST00000085133.13 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr3_-_146476331 | 0.12 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr6_-_16898440 | 0.12 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr3_-_146475974 | 0.12 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr9_+_44309727 | 0.12 |
ENSMUST00000213268.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr5_+_115373895 | 0.12 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr11_+_23234644 | 0.11 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr6_-_52181393 | 0.11 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr7_-_84328553 | 0.11 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_+_177272297 | 0.11 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_-_7400690 | 0.11 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr16_-_95387444 | 0.10 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor |
| chr3_+_84859453 | 0.10 |
ENSMUST00000029727.8
|
Fbxw7
|
F-box and WD-40 domain protein 7 |
| chr1_+_177273226 | 0.10 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr11_+_90078485 | 0.10 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr11_+_120123727 | 0.09 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr1_+_177272215 | 0.09 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr13_-_103901010 | 0.09 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr5_-_23821523 | 0.09 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr15_-_8740218 | 0.09 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_-_25147139 | 0.08 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr5_+_38233901 | 0.08 |
ENSMUST00000146864.6
|
Stx18
|
syntaxin 18 |
| chr10_+_87926932 | 0.08 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr2_+_72128239 | 0.07 |
ENSMUST00000144111.2
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr4_-_82768958 | 0.07 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr15_-_77037972 | 0.07 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrM_+_7779 | 0.07 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr12_-_55033113 | 0.07 |
ENSMUST00000038926.13
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr6_-_52141796 | 0.07 |
ENSMUST00000014848.11
|
Hoxa2
|
homeobox A2 |
| chr18_-_66155651 | 0.07 |
ENSMUST00000143990.2
|
Lman1
|
lectin, mannose-binding, 1 |
| chr4_+_11579648 | 0.07 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr17_+_34258411 | 0.06 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr4_+_128582519 | 0.06 |
ENSMUST00000106080.8
|
Phc2
|
polyhomeotic 2 |
| chr8_-_68427217 | 0.06 |
ENSMUST00000098696.10
ENSMUST00000038959.16 ENSMUST00000093469.11 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr12_-_55033130 | 0.06 |
ENSMUST00000173433.8
ENSMUST00000173803.2 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr3_+_68479578 | 0.06 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr11_+_93935021 | 0.06 |
ENSMUST00000075695.13
ENSMUST00000092777.11 |
Spag9
|
sperm associated antigen 9 |
| chr11_+_23256883 | 0.06 |
ENSMUST00000180046.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_+_93935066 | 0.06 |
ENSMUST00000103168.10
|
Spag9
|
sperm associated antigen 9 |
| chr3_-_37180093 | 0.06 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr2_-_6889783 | 0.06 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr4_-_92079986 | 0.06 |
ENSMUST00000123179.2
|
Gm12666
|
predicted gene 12666 |
| chr6_-_51443602 | 0.06 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr13_+_44994167 | 0.05 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr11_+_23256909 | 0.05 |
ENSMUST00000137823.8
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr19_+_26728339 | 0.05 |
ENSMUST00000175953.3
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr16_-_56533179 | 0.05 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr5_+_35146727 | 0.05 |
ENSMUST00000114284.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr10_+_18345706 | 0.05 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr12_-_34578842 | 0.05 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr5_-_123127148 | 0.05 |
ENSMUST00000046073.16
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr14_-_78866714 | 0.05 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr11_-_30218167 | 0.04 |
ENSMUST00000006629.14
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr1_+_6800275 | 0.04 |
ENSMUST00000043578.13
ENSMUST00000139756.8 ENSMUST00000131467.8 ENSMUST00000150761.8 ENSMUST00000151281.8 |
St18
|
suppression of tumorigenicity 18 |
| chr1_-_13061333 | 0.04 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr11_+_93935156 | 0.04 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr11_+_31823096 | 0.04 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr19_+_8861096 | 0.04 |
ENSMUST00000187504.7
|
Lbhd1
|
LBH domain containing 1 |
| chr5_+_35146880 | 0.04 |
ENSMUST00000114285.8
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_+_5724305 | 0.04 |
ENSMUST00000108380.2
|
Fam110b
|
family with sequence similarity 110, member B |
| chr8_-_47805383 | 0.04 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr9_+_118307412 | 0.04 |
ENSMUST00000035020.15
|
Eomes
|
eomesodermin |
| chr4_-_120672900 | 0.04 |
ENSMUST00000120779.8
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr3_-_137837117 | 0.04 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr6_+_29859660 | 0.04 |
ENSMUST00000128927.9
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_+_42459772 | 0.04 |
ENSMUST00000105497.8
ENSMUST00000144806.8 |
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr10_-_116385007 | 0.04 |
ENSMUST00000164088.8
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr6_+_29859685 | 0.04 |
ENSMUST00000134438.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr7_+_51537645 | 0.04 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr18_+_69652880 | 0.03 |
ENSMUST00000200813.4
|
Tcf4
|
transcription factor 4 |
| chr2_+_109522781 | 0.03 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr5_-_134485430 | 0.03 |
ENSMUST00000200944.4
ENSMUST00000202280.4 ENSMUST00000074114.12 ENSMUST00000100654.10 ENSMUST00000100650.10 ENSMUST00000111245.9 ENSMUST00000202554.4 ENSMUST00000073161.12 ENSMUST00000100652.10 ENSMUST00000171794.9 ENSMUST00000167084.9 |
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chr11_+_93934940 | 0.03 |
ENSMUST00000132079.8
|
Spag9
|
sperm associated antigen 9 |
| chr9_-_71803354 | 0.03 |
ENSMUST00000184448.8
|
Tcf12
|
transcription factor 12 |
| chr6_+_106095726 | 0.03 |
ENSMUST00000113258.8
ENSMUST00000079416.6 |
Cntn4
|
contactin 4 |
| chr18_+_35932894 | 0.03 |
ENSMUST00000236484.2
ENSMUST00000236550.2 ENSMUST00000235919.2 |
Ube2d2a
Gm50371
|
ubiquitin-conjugating enzyme E2D 2A predicted gene, 50371 |
| chr15_-_77037756 | 0.03 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr8_-_68270936 | 0.03 |
ENSMUST00000120071.8
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_+_120307390 | 0.03 |
ENSMUST00000110716.9
ENSMUST00000028748.14 ENSMUST00000090028.13 ENSMUST00000110719.4 |
Capn3
|
calpain 3 |
| chr6_-_13839914 | 0.03 |
ENSMUST00000060442.14
|
Gpr85
|
G protein-coupled receptor 85 |
| chr2_-_168048795 | 0.03 |
ENSMUST00000057793.11
|
Adnp
|
activity-dependent neuroprotective protein |
| chr2_+_20742115 | 0.03 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr19_-_46033353 | 0.03 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr12_-_101784727 | 0.02 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr10_-_6930376 | 0.02 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr13_+_83723255 | 0.02 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_7400780 | 0.02 |
ENSMUST00000002176.13
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr15_+_21111428 | 0.02 |
ENSMUST00000075132.8
|
Cdh12
|
cadherin 12 |
| chr9_+_32027335 | 0.02 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr10_+_127256192 | 0.02 |
ENSMUST00000171434.8
|
R3hdm2
|
R3H domain containing 2 |
| chr2_+_26481435 | 0.02 |
ENSMUST00000152988.9
ENSMUST00000149789.2 |
Egfl7
|
EGF-like domain 7 |
| chr9_-_71070506 | 0.02 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr7_-_37472979 | 0.02 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr14_-_121976273 | 0.02 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr18_-_84104507 | 0.02 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr6_+_15185399 | 0.02 |
ENSMUST00000115474.8
ENSMUST00000115472.8 ENSMUST00000031545.14 |
Foxp2
|
forkhead box P2 |
| chr15_-_77037926 | 0.02 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_100497863 | 0.02 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr11_+_102495189 | 0.02 |
ENSMUST00000057893.7
|
Fzd2
|
frizzled class receptor 2 |
| chr18_-_84104574 | 0.02 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr12_+_38833501 | 0.02 |
ENSMUST00000159334.8
|
Etv1
|
ets variant 1 |
| chr16_-_95260104 | 0.02 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr9_-_40257586 | 0.02 |
ENSMUST00000121357.8
|
Gramd1b
|
GRAM domain containing 1B |
| chr4_-_58499398 | 0.02 |
ENSMUST00000107570.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr9_-_13245834 | 0.02 |
ENSMUST00000110582.4
|
Jrkl
|
Jrk-like |
| chr13_+_16189041 | 0.02 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chr18_+_42669322 | 0.02 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr5_+_149601688 | 0.02 |
ENSMUST00000100404.6
|
B3glct
|
beta-3-glucosyltransferase |
| chr15_-_8739893 | 0.02 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_+_15196950 | 0.02 |
ENSMUST00000140557.8
ENSMUST00000131414.8 ENSMUST00000115469.8 |
Foxp2
|
forkhead box P2 |
| chr11_+_96214078 | 0.02 |
ENSMUST00000093944.10
|
Hoxb3
|
homeobox B3 |
| chr12_+_38833454 | 0.02 |
ENSMUST00000161980.8
ENSMUST00000160701.8 |
Etv1
|
ets variant 1 |
| chr3_-_9029097 | 0.02 |
ENSMUST00000091354.12
ENSMUST00000094381.11 |
Tpd52
|
tumor protein D52 |
| chr11_-_30148230 | 0.02 |
ENSMUST00000102838.10
|
Sptbn1
|
spectrin beta, non-erythrocytic 1 |
| chr18_-_81029986 | 0.02 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr4_-_110149916 | 0.02 |
ENSMUST00000106601.8
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr5_+_90666791 | 0.02 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr19_-_18978463 | 0.01 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr8_+_79755194 | 0.01 |
ENSMUST00000119254.8
ENSMUST00000238669.2 |
Zfp827
|
zinc finger protein 827 |
| chr4_-_14621805 | 0.01 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr11_+_116547932 | 0.01 |
ENSMUST00000116318.3
|
Prcd
|
photoreceptor disc component |
| chr2_-_168609110 | 0.01 |
ENSMUST00000029061.12
ENSMUST00000103074.2 |
Sall4
|
spalt like transcription factor 4 |
| chrX_-_87159237 | 0.01 |
ENSMUST00000113966.8
ENSMUST00000113964.2 |
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr17_+_3532455 | 0.01 |
ENSMUST00000227604.2
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr7_+_90739904 | 0.01 |
ENSMUST00000107196.10
ENSMUST00000074273.10 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr11_-_99265721 | 0.01 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr2_+_101455079 | 0.01 |
ENSMUST00000111227.2
|
Rag2
|
recombination activating gene 2 |
| chr18_+_23548455 | 0.01 |
ENSMUST00000115832.4
|
Dtna
|
dystrobrevin alpha |
| chr2_-_136958544 | 0.01 |
ENSMUST00000028735.8
|
Jag1
|
jagged 1 |
| chr13_-_103042554 | 0.01 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_187730018 | 0.01 |
ENSMUST00000027906.13
|
Esrrg
|
estrogen-related receptor gamma |
| chr4_-_35845204 | 0.01 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr3_+_129974531 | 0.01 |
ENSMUST00000080335.11
ENSMUST00000106353.2 |
Col25a1
|
collagen, type XXV, alpha 1 |
| chr9_-_60048503 | 0.01 |
ENSMUST00000034829.6
|
Thsd4
|
thrombospondin, type I, domain containing 4 |
| chr8_-_46747629 | 0.01 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor |
| chr12_+_29578354 | 0.01 |
ENSMUST00000218583.2
ENSMUST00000049784.17 |
Myt1l
|
myelin transcription factor 1-like |
| chr14_-_52248324 | 0.01 |
ENSMUST00000226964.2
|
Zfp219
|
zinc finger protein 219 |
| chr18_+_82929037 | 0.01 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr4_-_110144676 | 0.01 |
ENSMUST00000106598.8
ENSMUST00000102723.11 ENSMUST00000153906.2 |
Elavl4
|
ELAV like RNA binding protein 4 |
| chr18_-_14105670 | 0.01 |
ENSMUST00000025288.9
|
Zfp521
|
zinc finger protein 521 |
| chr7_-_115637970 | 0.01 |
ENSMUST00000166877.8
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_+_187730032 | 0.01 |
ENSMUST00000110938.2
|
Esrrg
|
estrogen-related receptor gamma |
| chr17_-_59298338 | 0.01 |
ENSMUST00000025064.14
|
Pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr5_-_86437119 | 0.01 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr6_-_138404076 | 0.01 |
ENSMUST00000203435.3
|
Lmo3
|
LIM domain only 3 |
| chr4_-_14621497 | 0.01 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr7_-_115459082 | 0.01 |
ENSMUST00000206123.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr15_+_28203872 | 0.01 |
ENSMUST00000067048.8
|
Dnah5
|
dynein, axonemal, heavy chain 5 |
| chr10_-_20600442 | 0.01 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr3_+_29568055 | 0.01 |
ENSMUST00000140288.2
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr19_+_55883924 | 0.01 |
ENSMUST00000111646.8
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr12_-_104439589 | 0.01 |
ENSMUST00000021513.6
|
Gsc
|
goosecoid homeobox |
| chr4_+_101365144 | 0.01 |
ENSMUST00000149047.8
ENSMUST00000106929.10 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr4_+_19280850 | 0.01 |
ENSMUST00000102999.2
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr7_+_91321500 | 0.01 |
ENSMUST00000238619.2
ENSMUST00000238467.2 |
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr14_-_48900192 | 0.01 |
ENSMUST00000122009.8
|
Otx2
|
orthodenticle homeobox 2 |
| chr2_-_168608949 | 0.01 |
ENSMUST00000075044.10
|
Sall4
|
spalt like transcription factor 4 |
| chr18_+_23548534 | 0.01 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr1_+_100025486 | 0.01 |
ENSMUST00000188735.2
|
Cntnap5b
|
contactin associated protein-like 5B |
| chr11_-_99412162 | 0.01 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr1_+_70764874 | 0.01 |
ENSMUST00000053922.12
ENSMUST00000161937.2 ENSMUST00000162182.2 |
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr1_-_126758369 | 0.01 |
ENSMUST00000112583.8
ENSMUST00000094609.10 |
Nckap5
|
NCK-associated protein 5 |
| chr6_-_138399896 | 0.01 |
ENSMUST00000161450.8
ENSMUST00000163024.8 ENSMUST00000162185.8 |
Lmo3
|
LIM domain only 3 |
| chr1_+_19173246 | 0.01 |
ENSMUST00000037294.8
|
Tfap2d
|
transcription factor AP-2, delta |
| chr3_+_55689921 | 0.01 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr5_+_20433169 | 0.01 |
ENSMUST00000197553.5
ENSMUST00000208219.2 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_-_13838423 | 0.01 |
ENSMUST00000115492.2
|
Gpr85
|
G protein-coupled receptor 85 |
| chr3_+_75982890 | 0.01 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr13_+_55547498 | 0.01 |
ENSMUST00000057167.9
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1 |
| chr5_+_13449276 | 0.01 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_81054992 | 0.01 |
ENSMUST00000068275.12
ENSMUST00000113494.9 |
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr6_+_34686373 | 0.01 |
ENSMUST00000115021.8
|
Cald1
|
caldesmon 1 |
| chr11_-_99412084 | 0.01 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.6 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.3 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0060574 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0035166 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.0 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |