Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
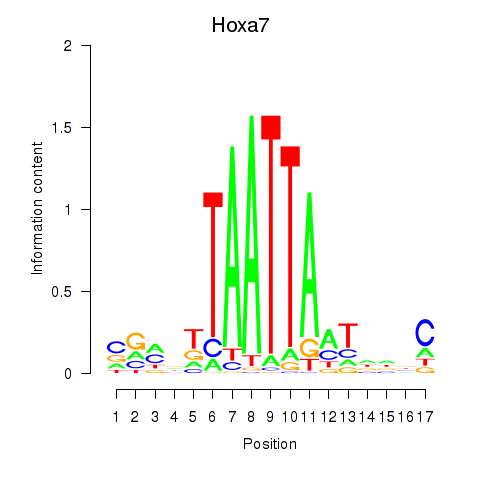
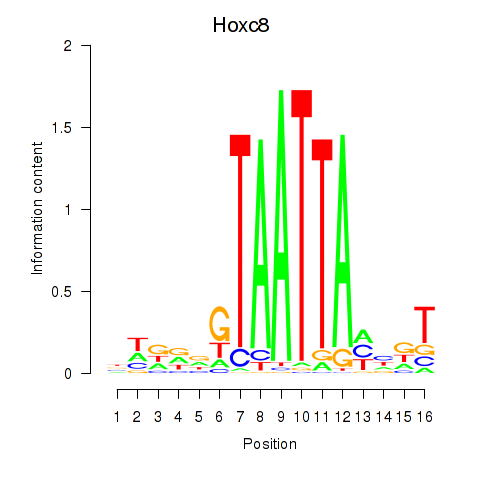
Results for Hoxa7_Hoxc8
Z-value: 0.45
Transcription factors associated with Hoxa7_Hoxc8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa7
|
ENSMUSG00000038236.9 | homeobox A7 |
|
Hoxc8
|
ENSMUSG00000001657.8 | homeobox C8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa7 | mm39_v1_chr6_-_52195663_52195696 | -0.85 | 6.6e-02 | Click! |
| Hoxc8 | mm39_v1_chr15_+_102898966_102899039 | 0.76 | 1.3e-01 | Click! |
Activity profile of Hoxa7_Hoxc8 motif
Sorted Z-values of Hoxa7_Hoxc8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_39932760 | 0.47 |
ENSMUST00000215956.3
|
Olfr981
|
olfactory receptor 981 |
| chr12_-_99529767 | 0.37 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr3_-_66204228 | 0.33 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_-_12829100 | 0.32 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr7_-_103113358 | 0.31 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chrM_+_9870 | 0.28 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr10_-_85847697 | 0.27 |
ENSMUST00000105304.2
ENSMUST00000061699.12 |
Bpifc
|
BPI fold containing family C |
| chr16_-_58620631 | 0.25 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173 |
| chr2_+_88470886 | 0.23 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr13_-_32967937 | 0.23 |
ENSMUST00000238977.3
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chrX_+_16485937 | 0.22 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A |
| chr9_+_40092216 | 0.21 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr9_+_38119661 | 0.19 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893 |
| chr2_+_83554770 | 0.19 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr5_-_116162415 | 0.19 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chrX_-_142716200 | 0.18 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr19_+_13339600 | 0.17 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467 |
| chrM_+_14138 | 0.16 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.16 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_+_100336003 | 0.16 |
ENSMUST00000133493.9
ENSMUST00000092730.5 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr4_+_8690398 | 0.16 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr10_+_127226180 | 0.15 |
ENSMUST00000077046.12
ENSMUST00000105250.9 |
R3hdm2
|
R3H domain containing 2 |
| chr9_-_58648826 | 0.14 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr18_+_32200781 | 0.14 |
ENSMUST00000025243.5
ENSMUST00000212675.2 |
Iws1
|
IWS1, SUPT6 interacting protein |
| chr13_+_22508759 | 0.14 |
ENSMUST00000226225.2
ENSMUST00000227017.2 |
Vmn1r197
|
vomeronasal 1 receptor 197 |
| chr9_+_19247753 | 0.13 |
ENSMUST00000215572.3
ENSMUST00000213344.2 |
Olfr845
|
olfactory receptor 845 |
| chr12_-_87742525 | 0.13 |
ENSMUST00000164517.3
|
Eif1ad19
|
eukaryotic translation initiation factor 1A domain containing 19 |
| chr7_+_84502761 | 0.13 |
ENSMUST00000217039.3
ENSMUST00000211582.2 |
Olfr291
|
olfactory receptor 291 |
| chr13_+_22656093 | 0.13 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr2_-_88994435 | 0.13 |
ENSMUST00000099793.3
|
Olfr1224-ps1
|
olfactory receptor 1224, pseudogene 1 |
| chr2_+_69727599 | 0.13 |
ENSMUST00000131553.2
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr11_-_73382303 | 0.12 |
ENSMUST00000119863.2
ENSMUST00000215358.2 ENSMUST00000214623.2 |
Olfr381
|
olfactory receptor 381 |
| chr2_-_45007407 | 0.12 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_29226886 | 0.12 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr15_-_66985760 | 0.12 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr7_-_102638531 | 0.11 |
ENSMUST00000215606.2
|
Olfr578
|
olfactory receptor 578 |
| chr19_-_53933052 | 0.11 |
ENSMUST00000135402.4
|
Bbip1
|
BBSome interacting protein 1 |
| chr10_-_128462616 | 0.10 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26 |
| chr5_-_23881353 | 0.09 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr5_-_62923463 | 0.09 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr14_+_80237691 | 0.09 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr11_+_29323618 | 0.08 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr19_+_53933271 | 0.08 |
ENSMUST00000025932.9
|
Shoc2
|
Shoc2, leucine rich repeat scaffold protein |
| chr9_+_74860133 | 0.08 |
ENSMUST00000215370.2
|
Fam214a
|
family with sequence similarity 214, member A |
| chr10_-_44024843 | 0.08 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr16_+_45044678 | 0.07 |
ENSMUST00000102802.10
ENSMUST00000063654.6 |
Btla
|
B and T lymphocyte associated |
| chr18_+_4920513 | 0.07 |
ENSMUST00000126977.8
|
Svil
|
supervillin |
| chr10_-_45346297 | 0.06 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chr13_-_53627110 | 0.06 |
ENSMUST00000021922.10
|
Msx2
|
msh homeobox 2 |
| chr10_-_75946790 | 0.06 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr13_+_22534534 | 0.06 |
ENSMUST00000226909.2
ENSMUST00000227167.2 ENSMUST00000226786.2 |
Vmn1r198
|
vomeronasal 1 receptor 198 |
| chrX_+_164953444 | 0.06 |
ENSMUST00000130880.9
ENSMUST00000056410.11 ENSMUST00000096252.10 ENSMUST00000087169.11 |
Gemin8
|
gem nuclear organelle associated protein 8 |
| chr9_-_119897358 | 0.06 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr11_-_73348284 | 0.06 |
ENSMUST00000121209.3
ENSMUST00000127789.3 |
Olfr380
|
olfactory receptor 380 |
| chr2_+_88479038 | 0.05 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr17_-_29226965 | 0.05 |
ENSMUST00000009138.13
ENSMUST00000119274.3 |
Stk38
|
serine/threonine kinase 38 |
| chr17_-_29226700 | 0.05 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr14_-_36641270 | 0.05 |
ENSMUST00000182797.8
|
Ccser2
|
coiled-coil serine rich 2 |
| chr9_-_119897328 | 0.05 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chrX_+_159551009 | 0.05 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr5_+_104350475 | 0.05 |
ENSMUST00000066708.7
|
Dmp1
|
dentin matrix protein 1 |
| chr4_-_114991478 | 0.05 |
ENSMUST00000106545.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr8_+_23901506 | 0.05 |
ENSMUST00000033952.8
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr6_-_137146708 | 0.04 |
ENSMUST00000117919.8
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr3_-_86455575 | 0.04 |
ENSMUST00000077524.4
|
Mab21l2
|
mab-21-like 2 |
| chr1_+_82294173 | 0.04 |
ENSMUST00000027322.14
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr4_+_108576846 | 0.04 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr16_+_13176238 | 0.03 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr2_+_87854404 | 0.03 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr11_+_73051228 | 0.03 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr6_-_122317484 | 0.03 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr16_-_19241884 | 0.02 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr4_+_145311759 | 0.02 |
ENSMUST00000119718.8
|
Zfp268
|
zinc finger protein 268 |
| chr17_-_45970238 | 0.02 |
ENSMUST00000120717.8
|
Capn11
|
calpain 11 |
| chr18_+_23886765 | 0.02 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr13_-_54713180 | 0.02 |
ENSMUST00000026989.15
|
4833439L19Rik
|
RIKEN cDNA 4833439L19 gene |
| chr10_+_97318223 | 0.02 |
ENSMUST00000163448.4
|
Dcn
|
decorin |
| chr16_-_45544960 | 0.02 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr18_-_10706701 | 0.01 |
ENSMUST00000002549.9
ENSMUST00000117726.9 ENSMUST00000117828.9 |
Abhd3
|
abhydrolase domain containing 3 |
| chr1_-_134883577 | 0.01 |
ENSMUST00000168381.8
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr4_-_43710231 | 0.01 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr2_-_86016027 | 0.01 |
ENSMUST00000215138.3
|
Olfr52
|
olfactory receptor 52 |
| chr19_-_38807600 | 0.01 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr7_-_103191924 | 0.01 |
ENSMUST00000214269.3
|
Olfr612
|
olfactory receptor 612 |
| chr6_-_30693675 | 0.01 |
ENSMUST00000169422.8
ENSMUST00000115131.8 ENSMUST00000115130.3 ENSMUST00000031810.15 |
Cep41
|
centrosomal protein 41 |
| chr10_-_129630735 | 0.01 |
ENSMUST00000217283.2
ENSMUST00000214206.2 ENSMUST00000214878.2 |
Olfr810
|
olfactory receptor 810 |
| chr16_-_75706161 | 0.01 |
ENSMUST00000114239.9
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr13_-_23041731 | 0.01 |
ENSMUST00000228645.2
|
Vmn1r211
|
vomeronasal 1 receptor 211 |
| chr7_-_25112256 | 0.01 |
ENSMUST00000200880.4
ENSMUST00000074040.4 |
Cxcl17
|
chemokine (C-X-C motif) ligand 17 |
| chr6_+_37847721 | 0.01 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr1_+_24216691 | 0.01 |
ENSMUST00000054588.15
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr2_-_85632888 | 0.01 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr16_-_88303859 | 0.00 |
ENSMUST00000069549.3
|
Cldn17
|
claudin 17 |
| chr5_-_99091681 | 0.00 |
ENSMUST00000162619.8
ENSMUST00000162147.6 |
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_-_134883645 | 0.00 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr11_-_73217633 | 0.00 |
ENSMUST00000134079.2
|
Aspa
|
aspartoacylase |
| chr7_-_46316767 | 0.00 |
ENSMUST00000168335.3
ENSMUST00000107669.9 |
Tph1
|
tryptophan hydroxylase 1 |
| chr15_-_82678490 | 0.00 |
ENSMUST00000006094.6
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr8_-_49008305 | 0.00 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr19_+_5524701 | 0.00 |
ENSMUST00000165485.8
ENSMUST00000166253.8 ENSMUST00000167371.8 ENSMUST00000167855.8 ENSMUST00000070118.14 |
Efemp2
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 2 |
| chr6_+_30541581 | 0.00 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr18_+_86729184 | 0.00 |
ENSMUST00000068423.10
|
Cbln2
|
cerebellin 2 precursor protein |
| chr6_-_41752111 | 0.00 |
ENSMUST00000214976.3
|
Olfr459
|
olfactory receptor 459 |
| chr16_-_93726399 | 0.00 |
ENSMUST00000177648.8
ENSMUST00000142083.2 |
Cldn14
|
claudin 14 |
| chr7_+_54485336 | 0.00 |
ENSMUST00000082373.8
|
Luzp2
|
leucine zipper protein 2 |
| chr5_+_29400981 | 0.00 |
ENSMUST00000160888.8
ENSMUST00000159272.8 ENSMUST00000001247.12 ENSMUST00000161398.8 ENSMUST00000160246.8 |
Rnf32
|
ring finger protein 32 |
| chr2_+_111327525 | 0.00 |
ENSMUST00000121345.4
|
Olfr1291-ps1
|
olfactory receptor 1291, pseudogene 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa7_Hoxc8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:0010793 | regulation of histone H3-K36 methylation(GO:0000414) regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0045275 | respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |